Using a Virtual Environment with Jupyter Notebook¶
The VACC Open OnDemand instance offers several choices of Python to choose from. The Anaconda distribution(s) with over 250 installed packages will cover most common scenarios except some specialized packages, e.g., Torch, TensorFlow.
This page gives a step-by-step guide to installing a particular set of
Python packages in a Conda virtual environment and then setting that
virtual environment up so it can be used with the Jupyter Notebook
application. Similarly, a virtual Python environment that was created
with either virtualenv or venv can be accessed in Jupyter Notebook
via a kernel.
Create a Conda virtual environment and install the kernel¶
These instructions assume that you are starting with a ‘clean slate’,
by which we mean there are no active Conda environments. This also
assumes that you want to create the virtual environment in the
standard path ~/.conda/envs. If you want to save it elsewhere, see
the official
documentation
for details on setting a --prefix. All of the commands that follow
are preceded by a $ prompt. If you copy and paste, do not copy the
prompt. Lines that start with a # are comments that simply provide
information. To be extra safe, we deactivate any Conda
environments. If you don’t have an active Conda environment, running
the deactivate command is harmless and will simply tell you that
The command: conda could not be found. This is a safety measure for
those for whom it is found.
$ conda deactivate
The first thing we want to do is insure that we have only the module(s) needed for our project, which is biology for this example. To do that we first remove all loaded modules, then load the one(s) we need.
# Clear modules.md
$ module purge
# Load the needed modules.md
$ module load python3.12-anaconda/2024.06-1
The next step is to source the conda.sh file. This will avoid
having to run conda init which would “permanently” add the Conda
setup commands to your ~/.bashrc file.
$ source ${ANACONDA_ROOT}/etc/profile.d/conda.sh
Next, we create the environment, making sure to specify the Python
version explicitly so that it matches one of the available options in
Open OnDemand. In this example, we name our environment
my_bio_env. You can substitute any name that is appropriate for your
environment. Additionally, note that the name given to the kernel does
not have to match the name of the conda environment. We use the same
name in our example below for simplicity.
$ conda create --name my_bio_env python=3.12
# Enter 'y' when asked: Proceed ([y]/n)?
$ conda activate my_bio_env
# Use the '-c' option to specify a channel if any of your
# packages need to be downloaded from a particular site
$ conda install -c conda-forge pandas numpy pySlim scikit-allel msprime
# Enter 'y' again, when asked to proceed
# Install ipykernel so that you can access your environment from
# the Jupyter Notebook menu (use -y to answer yes automatically)
$ conda install -y ipykernel
# Installing the kernel
$ python -m ipykernel install --user --name=my_bio_env
# Deactivate when finished
$ conda deactivate
Using the new kernel with Jupyter Notebook¶
Go to VACC Open OnDemand, bring up
the JupyterLab / Jupyter Notebook form and select Jupyter Notebook
mode. Under the Python Version field, be sure to select a version that
matches the Python version used when you created your environment. In
this case, python3.12-anaconda/2024.06-1 would be the appropriate
version to choose.
Once your Jupyter notebook has started, you should see my_bio_env
(or the name given to your virtual environment) listed as an available
kernel under the ‘New’ pull down menu.
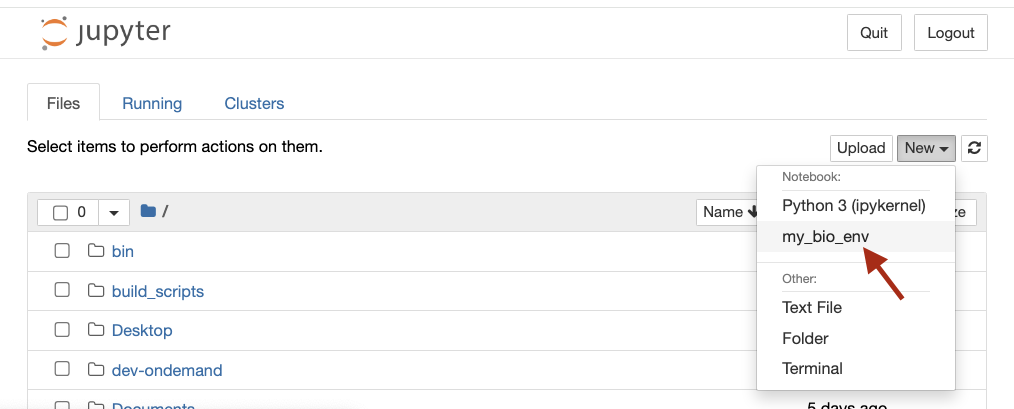 Screenshot of Jupyter Notebook showing the installed kernel as an option in the "New" dropdown menu.
Screenshot of Jupyter Notebook showing the installed kernel as an option in the "New" dropdown menu.
Using the new Conda environment from a terminal or in a SLURM batch job¶
To use the new Conda virtual environment, in this case named
my_bio_env, from within a terminal, enter the following commands:
$ module load python3.12-anaconda/2024.06-1
$ source ${ANACONDA_ROOT}/etc/profile.d/conda.sh
$ conda activate my_bio_env
If you want to run a batch job that uses the Conda environment, copy
the above commands into your batch script. Do not include the $ at
the beginning of each command.