Chapter 6 Data Wrangling - Part 1
Packages Required:
* lubridate - a package for working with dates
* ggplot2 - a graphics package
6.1 Introduction
Let’s now wander into the land of Tauntauns and Tauntaun conservation and management. We’ll assume that you are a new scientist working for a conservation organization in Vermont, USA, and are charged with assessing the population status of the Tauntauns. The harvest keeps the Tauntaun population from exploding, and the sale of harvest licenses financially supports the conservation of all other species in Vermont.

Figure 6.1: Retrieved on March 24 2015 from http://starwars.wikia.com/wiki/Tauntaun.
Recall that our imaginary population of Tauntauns here in Vermont is managed via a harvest. Let’s assume the previous biologist has left you with a dataset that contains records about each Tauntaun that has been harvested through time. In addition, there is another dataset that contains information about Tauntaun hunters. Your job is to take the harvest data records, which is full of information about dead animals, and figure out how well the living population is faring (in terms of population size and trend). Good luck! Your job also requires that you produce an annual report to the U.S. Fish and Wildlife Service, which supports your conservation agency through grants. This annual report is a simple summary of the harvest for the year, but needs to be clearly written.
You’ve heard that R is a great tool for data analysis and reporting – you’ve downloaded R and RStudio onto your new laptop and are eager to get started. Of course, you’ve read Chapters 1-4 of this primer, so have a general working knowledge of RStudio, functions, and objects. In Chapter 5 you created the Tauntaun project, and added the directories and files needed to start working.
Our mission now is to work with two of these files and get comfortable manipulating data in R, a process known as “data wrangling”. The two files of interest are TauntaunHarvest.csv and hunter.csv, which should be located in your datasets directory. In this chapter, our focus will be on importing datasets to R, learning functions for examining and cleaning the data, and exporting the “cleaned” datasets as new .RData files. Then, in Chapter 7, we’ll continue building our function repertoire for working with data . . . we’ll be merging datasets, including the spatial information we obtained from the web. In Chapter 8, we introduce a package called dplyr. This package has several functions for data wrangling, and allows a coding approach that keeps your code clean and concise. After all this data wrangling, were ready for some statistical analysis, covered in Chapter 9. Finally, we can write our Tautaun annual report as an Rmarkdown document, covered in Chapter 10. Let’s go.
6.2 Create a New Script
Our goal in this chapter is to clean up the two csv files that store harvest and hunter information, and to save the “clean” versions to a RData file. We’ll save the code in a new script so that you can re-use it in future years as new Tauntaun data come in.
Open up your Tautaun project by clicking on its .Rproj file. Then go to Files | New File | R script. RStudio’s script editor should display a new file called Untitled1.
Figure 6.2: Create a new R script in your Tauntaun project.
When you begin a new script, no matter what coding language, it is a very good idea to write notes about what the file is about. This is fairly standard practice for coders. The notes can description of what this file is about, the author of the file, and the date on which the file is created. These will be entered as “comments” - which start with a number symbol or hashtag (#). Remember that R does not interpret comments; the comments are there so that you, the coder, can what code is doing. Get in the practice of commenting liberally! Copy the following into your script:
# This file is used to check and edit the Tauntaun harvest datasets. The final datasets
# will be exported as .RData files called "harvest_clean.RData" and "hunter_clean.RData",
# which will be used in the next chapter.
# Script coded by <enter your name here>
# Date: <enter today's date>.
RStudio has a helper for adding hashtags quickly. Just type your lines, highlight them, and then go to the toolbar and select Code | Comment/Uncomment Lines, and RStudio will add the hashtags for you. (Note the keyboard shortcuts if you don’t like to use your mouse.)
Before we go any further, save this file as TauntaunDataPrep.R. Look for this file in your Files pane…if it’s not there, press the refresh button and you should then see it.
6.3 Read in the Harvest CSV file
In Chapter 5, you downloaded the two csv files into your datasets directory.
- TauntaunHarvest.csv
- hunter.csv
Now we’re ready to read them into R as new objects in the global environment with read.csv. Let’s have a look at this function’s arguments:
## function (file, header = TRUE, sep = ",", quote = "\"", dec = ".",
## fill = TRUE, comment.char = "", ...)
## NULLBefore we use this function, remember that read.csv is a wrapper for read.table. That means that the read.csv function actually uses the read.table function, but simplifies it for csv files (comma separated variables). The dots argument in the read.csv function allows us to pass in additional arguments that we can find for read.table. Let’s look at the arguments for read.table:
## function (file, header = FALSE, sep = "", quote = "\"'", dec = ".",
## numerals = c("allow.loss", "warn.loss", "no.loss"), row.names,
## col.names, as.is = !stringsAsFactors, na.strings = "NA",
## colClasses = NA, nrows = -1, skip = 0, check.names = TRUE,
## fill = !blank.lines.skip, strip.white = FALSE, blank.lines.skip = TRUE,
## comment.char = "#", allowEscapes = FALSE, flush = FALSE,
## stringsAsFactors = default.stringsAsFactors(), fileEncoding = "",
## encoding = "unknown", text, skipNul = FALSE)
## NULLThere are several arguments here, but one that will affect our Tauntaun data analysis is the argument, stringsAsFactors, which can be set to TRUE or FALSE. This argument pertains to columns in your CSV file that are characters (strings). The argument is asking whether such columns should be imported as characters (FALSE) or if they should be imported as factors (TRUE). We will use pass this read.table argument as a read.csv dots argument in the code below. Note that you don’t actually use the dots – just use the read.table argument names. Also note that the file name is enclosed with quotes, which tells R it is a character value as opposed to an existing object.
# read in the Tauntaun harvest dataset
harvest <- read.csv(file = "datasets/TauntaunHarvest.csv",
stringsAsFactors = TRUE)You could also use RStudio’s import wizard by clicking on the file itself in the Files tab. An import wizard will open up and lead you through the import steps.
Did you notice a new object in your Environment called harvest?
6.4 Looking at the Data
Now let’s take a look at the object called harvest. The str function is useful as a first step for a quick overview:
## 'data.frame': 13351 obs. of 12 variables:
## $ hunter.id: int 285 56 549 296 521 355 674 297 7 388 ...
## $ age : int 1 1 1 1 1 1 1 1 1 1 ...
## $ sex : Factor w/ 2 levels "f","m": 1 1 1 1 1 1 1 1 1 1 ...
## $ individul: int 1 2 3 4 5 6 7 8 9 10 ...
## $ species : Factor w/ 3 levels "tantaun","tauntaan",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ date : Factor w/ 920 levels "10/1/1901","10/1/1902",..: 291 351 791 631 441 91 551 251 401 661 ...
## $ town : Factor w/ 255 levels "ADDISON","ALBANY",..: 196 10 92 192 241 152 8 202 136 32 ...
## $ length : num 277 312 275 267 272 ...
## $ weight : num 699 718 551 700 613 ...
## $ method : Factor w/ 3 levels "bow","lightsaber",..: 2 3 1 3 1 2 2 1 2 2 ...
## $ color : Factor w/ 3 levels "gray","grey",..: 3 1 3 1 1 2 3 1 3 3 ...
## $ fur : Factor w/ 3 levels "long","medium",..: 1 1 1 1 1 3 3 1 2 1 ...Notice that the read.csv function returns a data frame. Here, we can see the data frame has 13351 observations (rows) and 12 variables (columns). That’s a lot of Tauntauns!
By now, you should be able to make sense of each and every column (identified by the $ in the structure output). You might recall from a previous chapter that data frames are a special kind of list made up of multiple vectors that are all the same length. Each element of a list (or each vector) can be identified with a dollar sign, which can be used to extract a given list element (in this case, a particular column).
Note that R thinks that 3 of the columns contain integer data types, 7 of the columns contain factor data types, and 2 of the columns contain numeric data types. This might not be what we want, so we’ll need to work through each column and fix things as needed.
It’s helpful to look at the dataset directly to get a better feel for it. Let’s scan our data with the head and tail functions. These related functions both have an argument named x (which is the dataset) and an argument named n (which identifies the number of lines to display):
## hunter.id age sex individul species date town length weight method color fur
## 1 285 1 f 1 tauntaun 10/8/1901 STANNARD 277.10 699.3044 lightsaber white long
## 2 56 1 f 2 tauntaun 11/13/1901 BALTIMORE 311.86 718.1022 muzzle gray long
## 3 549 1 f 3 tauntaun 12/26/1901 HANCOCK 274.84 550.6420 bow white long
## 4 296 1 f 4 tauntaun 12/11/1901 ST. ALBANS TOWN 266.70 699.5951 muzzle gray long
## 5 521 1 f 5 tauntaun 11/21/1901 WHEELOCK 271.84 613.3350 bow gray longTo save space, we may not show the helpfile function call from this point forward, but please use
helpliberally, especially when you are using a function for the first time.
Now let’s use the tail function to look at the last 4 records:
## hunter.id age sex individul species date town length weight method color fur
## 13348 504 5 m 13348 tauntaun 11/3/1910 STRATTON 425.65 1069.7622 muzzle gray short
## 13349 290 5 m 13349 tauntaun 10/19/1910 AVERYS GORE 442.25 1099.0944 lightsaber gray short
## 13350 681 5 m 13350 tauntaun 10/15/1910 SWANTON 392.91 951.8551 muzzle gray long
## 13351 634 5 m 13351 tauntaun 10/4/1910 ALBANY 448.35 1155.6545 lightsaber gray longYou can also look at the data in a “spreadsheet” type of view. Let’s use the View function to take a look at the entire dataset (note the capital “V”). This function invokes a spreadsheet-style data viewer, and the data are displayed as a new tab in the Scripts pane, so now you’ll have two tabs in the the Scripts pane (harvest, and TauntaunDataPrep.R). You can easily navigate between the two by just clicking on them. Note: clicking on the grid icon associated with the object called harvest in RStudio’s Environment tab will also invoke the View function.
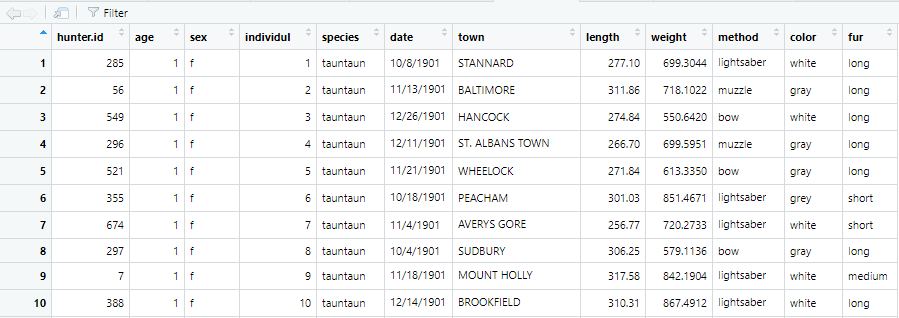
Figure 6.3: the View function will open the dataset in a new tab in the Script editor.
R shows the first 1000 lines only, although more lines are present.
Let’s find out more about this dataset’s dimensions. We first want to count the rows and columns with the functions nrow and ncol.
## [1] 13351## [1] 12There is another command to query the dimensions of the dataset that will report these values with a single command:
## [1] 13351 12The catch with dim is that we need to remember that R always indexes and reports rows first, then columns. Remember Russell Crowe? (How about Ray Charles, Rosemary Cloooney, or Roberto Clemente?)
Another function that is often used to learn about an object is the length function:
## [1] 12This is a very long dataset with thousands of rows, so we can be certain that 12 is not referring to the number of rows. Using the length function on a data frame object returns the number of columns. Why is this? Once again, it has to do with the fact that data frames are a special kind of a list. Here, length is giving us the the number of objects within the data frame list, which is 12 (the dataframe is list containing 12 objects that are all vectors of size 13351).
Did you read the helpfiles on these functions?
6.5 Summarizing Data
We’ve seen that the str function summarizes the structure of the object. It prints the object’s class, dimensions, element names, element classes (data types), and provides a 10-value peek at the contents of each element. But it doesn’t tell us much about the actual contents of objects. There are a couple of reasonable approaches to investigating what’s inside a new data frame (or any new object in R). The first way is to get a summary of each and every column with the summary function.
## hunter.id age sex individul species date town length weight
## Min. : 1.0 Min. :1.000 f:6161 Min. : 1 tantaun : 14 10/5/1909 : 42 DUMMERSTON : 69 Min. : 228.8 Min. : 345.9
## 1st Qu.:176.0 1st Qu.:1.000 m:7190 1st Qu.: 3338 tauntaan: 20 10/14/1910: 41 LONDONDERRY: 69 1st Qu.: 301.6 1st Qu.: 748.7
## Median :349.0 Median :2.000 Median : 6676 tauntaun:13317 10/22/1908: 38 SWANTON : 68 Median : 343.4 Median : 871.2
## Mean :351.2 Mean :1.953 Mean : 6676 10/4/1910 : 38 CHESTER : 67 Mean : 355.1 Mean : 883.2
## 3rd Qu.:528.0 3rd Qu.:3.000 3rd Qu.:10014 10/10/1910: 37 TINMOUTH : 67 3rd Qu.: 403.0 3rd Qu.:1012.4
## Max. :700.0 Max. :5.000 Max. :13351 10/15/1910: 35 WOODBURY : 67 Max. :4591.8 Max. :1521.2
## (Other) :13120 (Other) :12944
## method color fur
## bow :3168 gray :8193 long :4604
## lightsaber:6454 grey :1306 medium:4334
## muzzle :3217 white:3852 short :4413
## NA's : 512
##
##
## You can see that R provides a summary column by column. Our dataset features a mix of datatypes. Notice that the numerical variables (hunter.id, age, individul, weight, length) are summarized differently than categorical variables (sex, species, date, town, method, color, fur). For numeric columns, summary returns the min, max, mean, 1st quartile (the 25th percentile), median (the 50th percentile), and 3rd quartile (the 75th percentile). For character columns, summary returns a count of each unique record within the column.
The summary function is designed to summarize the data in an object, not the object itself. You’ll probably notice a few spelling errors in the data (like the column name individul; it was entered by volunteers, so we can’t really complain), and there is at least one column (method) with NA values, which indicate missing data. In addition to fixing typos, we need to make sure the variables are of the correct data type or our analyses might be faulty. As painful as it may sound, it’s critical to walk through each and every column and ensure things are in order.
We’ll be using several functions along the way in our data wrangling effort:
- the trusty
strfunction - the
uniquefunction - the
histfunction - the
boxplotfunction - the
histogramfunction from the package, lattice - the
tablefunction - the
namesfunction - the
quantilefunction - descriptive functions, such as
min,max,range,mean,sd - coercion functions, such as
as.character,as.Date - and more!
Note: Our goal here is to introduce these functions to you as a way to inspect your data. In the name of keeping this chapter as short as possible, we’ll use the
strfunction plus one additional function to explore each column so you can see what the function does. You will probably want to be more thorough when working with real data!
Here we go.
6.6 Extracting a Column
There are a variety of ways to extract columns in a dataframe. Here, we will show you some of the most common extractor functions that are used to do this, including brackets, double brackets, and the dollar sign.
Suppose we want to examine the structure of the first column of our data frame, which stores the variable hunter.id. Remember that a data frame is a list that is made up of many vectors (12 in our case), each of the same length. There are several ways to get the structure of the first column only, and we provide 5 examples below. Pay attention to what sort of object is returned . . . they vary depending on method.
## int [1:13351] 285 56 549 296 521 355 674 297 7 388 ...# option 2: single bracket extraction by element name; this returns a data frame
str(harvest['hunter.id'])## 'data.frame': 13351 obs. of 1 variable:
## $ hunter.id: int 285 56 549 296 521 355 674 297 7 388 ...## 'data.frame': 13351 obs. of 1 variable:
## $ hunter.id: int 285 56 549 296 521 355 674 297 7 388 ...## int [1:13351] 285 56 549 296 521 355 674 297 7 388 ...## int [1:13351] 285 56 549 296 521 355 674 297 7 388 ...The take home point is that the output is differs depending on which method you used. Using the dollar-sign or double-bracket extractor returns a vector, while using single-bracket notation returns a data frame.
If you search the helpfiles on bracket notation (e.g., help(“[”)) you’ll see that the functions [ ], [[ ]], and $ are all “extractor functions”. You’ll also see that “The most important distinction between [, [[ and $ is that the [ can select more than one element whereas the other two select a single element.”
Since these are functions in their own right, they have arguments such as name, drop, exact, and value. The arguments are often dismissed in coding, but knowing they are there can be useful. R’s helpfile on extracting from a dataframe has more information.
For our purposes, we just need to inspect the data types and find a way of visualizing the data in an effort to sniff out potential errors, so we’ll use the $ extractor, which returns a vector (the column itself).
6.6.1 hunter.id = the unique hunter identification number
Our first column is called hunter.id. Each Tauntaun hunter is assigned a unique identifier (an integer), which allows us to determine how many Tauntauns were harvested per hunter. First, let’s confirm the structure:
## int [1:13351] 285 56 549 296 521 355 674 297 7 388 ...You can see that the id is stored as an integer. We can count the total number of hunters by combining the length and unique functions. The unique function will return all of the entries that are unique in the column as a vector. Nesting this function within the length function will give us our hunter count:
## [1] 700So, 700 hunters have registered a harvested Tauntaun. (In case you were wondering, Han Solo is not among them).
6.6.2 age = the age of the harvested animal
The next column in our dataset is age, which refers to the age of the harvested animal. Let’s look at the structure for this column and extract the unique entries using the unique function again. The unique function doesn’t sort the values within the column; it just reports them in the order they are encountered.
## int [1:13351] 1 1 1 1 1 1 1 1 1 1 ...# notice the unique ages in the datasets range from 0-5, presented in the order in which they appear
unique(harvest$age)## [1] 1 2 3 4 5This, too, looks to be a correct interpretation of the data within this column because all of the records are integers. It appears that the dataset consists of Tauntauns that range in age between 1 and 5 years old. Tauntauns drop dead on their 6th birthdays, so there are no 6 year olds in the dataset.
6.6.3 sex = the sex of the harvested animal
Let’s move on to the next column, which gives the sex of the harvested animal. You’ll see that R is treating this variable as a factor. With factors, the table function is a quick way to summarize counts.
## Factor w/ 2 levels "f","m": 1 1 1 1 1 1 1 1 1 1 ...## [1] "f" "m"##
## f m
## 6161 7190We briefly touched on “factors” in Chapter 4, and here is your first look at a factor data type. The str output shows that sex is a factor with 2 levels: “f” and “m”. These are coded as 1’s and 2’s, but how do you know which one is which? The output gives you a hint because the levels are reported in the order in which R encounters them (so 1 = “f”). For a factor like sex, knowing which numerical value corresponds with which character value will not make life easier. The exception to this rule is when the levels themselves are ordered, such as ‘small’, ‘medium’, and ‘large’. In that case, it’s important to understand the levels are ordered. This is not the case here. Later in the chapter we’ll look at the column, fur length, which is an ordered factor.
Using the factor system saves storage space when saving R objects with the save function, plus it conserves valuable memory during the R session. Also, for some statistical functions and graphic functions, the data must be stored as factors for the function to work as intended. On the other hand, factors can be cumbersome at first, so you may find yourself temporarily disabling them when reading in csv files using the argument stringsAsFactors = FALSE, or converting them to characters if needed.
As an aside, let’s quickly see how we can store a table as a type of object.
# if the results of the table function are stored in R, the object type is "table"
gender <- table(harvest$sex)
# look at the gender object
gender##
## f m
## 6161 7190## 'table' int [1:2(1d)] 6161 7190
## - attr(*, "dimnames")=List of 1
## ..$ : chr [1:2] "f" "m"The object, gender is of class table. We didn’t introduce this class in Chapter 4, but it is useful to know about, particularly when you need to summarize frequencies.
6.6.4 individual = the unique identifier of each harvested animal
The next column to examine is individul (a typo for individual), which identifies each harvested Tauntaun with a unique number. Identifying records in this manner (i.e. an auto-number approach) is typical of a database approach in data management.
We can pull the names of the columns of our harvest dataframe with the names function. This function is used to get or set the names of an object.
## [1] "hunter.id" "age" "sex" "individul" "species" "date" "town" "length" "weight" "method" "color" "fur"When run on a data frame like this, names is equivalent to colnames. As we saw in Chapter 4, the names function can also be used to name elements in lists, vectors, and arrays. Used like we did here, the names function returns an attribute of the object, harvest, that is called names. Let’s take a look:
## [1] "hunter.id" "age" "sex" "individul" "species" "date" "town" "length" "weight" "method" "color" "fur"We can also use the names function (which returns a vector) to replace one or all of the column names. To use it as a replacement function, first we use names(harvest) to return a vector of names, and then identify the element within the vector by either number or name. In this case we want to replace the name of the 4th column, so we index the 4th value of our returned names vector:
# extract the fourth element from the names vector, and replace it with 'individual'
names(harvest)[4] <- 'individual'This works just fine, as long as individual will always be the fourth column in your dataframe. An equivalent way to do this is to extract the proper element by name, which is probably a bit safer. In the next bit of code, we use the vector from names(harvest), and then subset this vector with the brackets [ ]. Inside the brackets, we look for the element within names(harvest) whose value == ‘individul’ and then replace it with the proper spelling, like this:
# extract from the vector of names the element whose value == 'individul'
names(harvest)[names(harvest) == 'individul'] <- "individual"
# check the names
names(harvest)## [1] "hunter.id" "age" "sex" "individual" "species" "date" "town" "length" "weight" "method" "color"
## [12] "fur"This is the first time we’ve used the == notation, so let’s talk about it briefly. In Chapter 1, we mentioned that you can use the = symbol to assign values to an object (but then we quickly suggested that you use the assignment operator, <- instead). Since = can be used to assign objects, R uses the == notation as a test of whether two things are equal to each other. R will evaluate the objects on both sides of the == symbol, and then return TRUE if they are equal and FALSE if they are not. The code names(harvest) will first extract the names of our harvest dataset (a vector), while the code [names(harvest)==‘Individul’] asks whether each element is ‘Individul’. This returns a vector (behind the scenes of TRUEs and FALSEs), like this:
## [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSEWe’ve already corrected the spelling, which is why there are 12 FALSE values here. When combined together,
names(harvest)[names(harvest)==‘individul’] <- “individual”
will subset the names of the harvest dataframe by the offending column (TRUE) and replace it with the correct name, individual.
6.6.5 species = the species harvested
Our next column is called species. Let’s take a look at its structure and count the unique entries with the table function again:
## Factor w/ 3 levels "tantaun","tauntaan",..: 3 3 3 3 3 3 3 3 3 3 ...##
## tantaun tauntaan tauntaun
## 14 20 13317R is interpreting this column as a factor with three levels, and you can see the first 10 records are all assigned a code of 3. Our table of Tauntaun harvest appears to either contain data for 3 species or it has 3 different spellings of Tauntaun. Believe it or not, the easiest way to fix the misspelled species names in this case is to replace the entire column with the correct spelling. Fear not, this is actually an extremely simple task. To do this, we index the column by name or index, and we assign it a single value. If we assign a single value to a column, R will recycle it as many times as necessary to fill the column.
# set all entries in the species column to Tauntaun
harvest['species'] <- 'Tauntaun'
# check the structure again
str(harvest$species)## chr [1:13351] "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" "Tauntaun" ...R has converted the species column from a factor to a character data type. This is fine for our purposes. As you can see, there are several ways to index from a data frame and several ways to replace these values.
6.6.6 date = the day of harvest
We’re on a roll. The next column is date, and like in most programming languages, date and time data types require special attention. First, let’s take a look to see how R is storing this value:
## Factor w/ 920 levels "10/1/1901","10/1/1902",..: 291 351 791 631 441 91 551 251 401 661 ...The date column is stored as factor…notice the quotes around the dates (e.g., 10/8/1901), which are followed by the numeric mapping of each date to a factor level. While this might be useful for some analyses, in our Tauntaun analysis we want this to be stored as a date. We briefly touched on the as.Date function to coerce values to dates. But let’s read the helpfile and dig into this topic more deeply.
The helpfiles show several arguments:
- x is the object to be converted.
- format = A character string. If not specified, it will try “%Y-%m-%d” then “%Y/%m/%d” on the first non-NA element, and give an error if neither works.
- origin = a Date object, or something which can be coerced by as.Date(origin, …) to such an object.
Hmmmmm. Well, at least x is easy to identify: harvest$date. We can see from our str output that the raw data look something along the lines of “10/1/1901”, or month/day/year. Year is written with 4 digits. The format argument is a character string, and it looks like %m-%d-%Y matches the syntax of our data. The option with a lower case y %m-%d-%y would be used if year was formatted as a two-digit code. To see other date formats, see strftime.
The catch here is that you must first convert the factor to character, and then convert the character to date. The reason for this should become clear in another minute or two. In both steps the result is stored, replacing the original data.
# first convert the data to characters
harvest$date <- as.character(harvest$date)
# then use the as.Date function to convert the factors to dates
harvest$date <- as.Date(harvest$date, format = "%m/%d/%Y")
# confirm the result
str(harvest$date)## Date[1:13351], format: "1901-10-08" "1901-11-13" "1901-12-26" "1901-12-11" "1901-11-21" "1901-10-18" "1901-11-04" "1901-10-04" "1901-11-18" "1901-12-14" ...That worked, and we should be able to summarize our harvests by months or years in the next chapter.
Dates and time are often stored as numbers in computers, and R is no different. Let’s quickly demonstrate this. Remember when we looked at Nelson Mandela’s birthday?
## [1] "1918-07-18"## [1] "Date"Let’s now convert the date to a number:
## [1] -18795-18795? Storing dates and times as numbers allows R to do arithmetic with dates. But this is also why converting directly from a factor (which is a number) directly to a date will cause problems.
If Nelson Mandela’s birthday is -18795, what is Day 0? Day 0 establishes the origin, and R’s default origin is 1970-01-01. Let’s double check the default origin:
## [1] "1970-01-01"## [1] 0In 1970, 8 tracks were blaring, the Beatles broke up, and floppy disks were introduced. Nelson Mandela was born -18795 days before Jan 1 1970. You can use the origin argument in the as.Date function to set the origin to any value you want; we’ve used R’s default.
By the way, although the time of harvest is not stored in this dataset, times are stored as fractions of days. For example, -18795.5 means day -18795 and 12 hours, 0 minutes, and 0 seconds.
As the Tauntaun biologist charged with analyzing data, you’d like to create four new columns, month, year, julian, day.of.season to help you summarize your harvest dates. Dates and times are tricky business - leap years, time zones, formatting, etc. are all knuckleballs that need to be considered.
We can take the “dummies” path and load library lubridate, one of the many excellent packages authored by Hadley Wickham and part of the tidyverse collection of R packages. Lubridate’s vignette has this to say:
Getting R to agree that your data contains the dates and times you think it does can be tricky. Lubridate simplifies that.
If you haven’t installed lubridate yet, do so now. Let’s load lubridate with the library function.
Make sure to skim through lubridate’s manual and at least read the list of functions that are available.
Our date column is already formatted as dates in R, but we can use lubridate’s month function to quickly create a new column called month.
# create the month column using lubridate's month function
harvest$month <- month(harvest$date)
# check the data frame and confirm it is present
head(harvest)## hunter.id age sex individual species date town length weight method color fur month
## 1 285 1 f 1 Tauntaun 1901-10-08 STANNARD 277.10 699.3044 lightsaber white long 10
## 2 56 1 f 2 Tauntaun 1901-11-13 BALTIMORE 311.86 718.1022 muzzle gray long 11
## 3 549 1 f 3 Tauntaun 1901-12-26 HANCOCK 274.84 550.6420 bow white long 12
## 4 296 1 f 4 Tauntaun 1901-12-11 ST. ALBANS TOWN 266.70 699.5951 muzzle gray long 12
## 5 521 1 f 5 Tauntaun 1901-11-21 WHEELOCK 271.84 613.3350 bow gray long 11
## 6 355 1 f 6 Tauntaun 1901-10-18 PEACHAM 301.03 851.4671 lightsaber grey short 10Month is stored as integers in column 13, which is fine.
You’ve just seen that you can create a new column column called month by typing in harvest$month, and then assigning a value to it. You could also have entered harvest[‘month’] <- month(harvest$date) to achieve the same result.
In the future we will be analyzing data by year, so to make that easier we can make a column called “year”. Lubridate’s year function can pull the year values from the date column:
# create a year column with and add values from lubridate's year function
harvest['year'] <- year(harvest[, 'date'])Let’s have a look:
## hunter.id age sex individual species date town length weight method color fur month year
## 1 285 1 f 1 Tauntaun 1901-10-08 STANNARD 277.10 699.3044 lightsaber white long 10 1901
## 2 56 1 f 2 Tauntaun 1901-11-13 BALTIMORE 311.86 718.1022 muzzle gray long 11 1901
## 3 549 1 f 3 Tauntaun 1901-12-26 HANCOCK 274.84 550.6420 bow white long 12 1901
## 4 296 1 f 4 Tauntaun 1901-12-11 ST. ALBANS TOWN 266.70 699.5951 muzzle gray long 12 1901
## 5 521 1 f 5 Tauntaun 1901-11-21 WHEELOCK 271.84 613.3350 bow gray long 11 1901
## 6 355 1 f 6 Tauntaun 1901-10-18 PEACHAM 301.03 851.4671 lightsaber grey short 10 1901Now suppose we want another column that indicates the day of the year in which the harvest occurred (called julian). In the Julian day system, Jan 1 is day 1 while Dec 31st is day 365 (unless it is a leap year). As the Tauntaun biologist, you think this information might be useful to estimate population size (after you’ve settled into your job, that is).
We can use lubridate’s yday function to convert our dates to “Julian” dates (the day of the year, with Jan 1 being day 1).
# create a new column in harvest called "julian"
harvest$julian <- yday(harvest$date)
# extract rows 1:5, columns "date", "month", "year", and "julian"
harvest[1:5, c("date", "month", "year", "julian")]## date month year julian
## 1 1901-10-08 10 1901 281
## 2 1901-11-13 11 1901 317
## 3 1901-12-26 12 1901 360
## 4 1901-12-11 12 1901 345
## 5 1901-11-21 11 1901 325You can see that animals are harvested in October, November, and December. You can also see the corresponding Julian dates. This is only the first five animals in our dataset, so let’s inspect all of the Julian data with a few functions that you may be familiar with:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 274.0 289.0 305.0 310.4 329.0 366.0## [1] 274 366## [1] 274## [1] 366## [1] 310.4176So, the average Julian harvest date was day 310.418 of the year, just before it starts getting cold in Vermont.
The Tauntaun harvest season always begins on October 1st (which is the UN’s International Day of Older Persons, and set in honor of the elder Obi-wan Kenobi). It would be useful to find the Julian date for October 1, and create a new column called day.of.season which returns a 1 for animals harvested on October 1, a 2 for animals harvested on October 2, a 5 for animals harvested on October 5th, etc. Let’s try that now. This will take a few steps because the Julian day for October 1 will differ depending on whether the year is a leap year or not. Let’s start by creating a vector of October 1st dates for all years of interest, and then converting them to Julian days:
## [1] 10 10 10 10 10 10 10 10 10 10## [1] 1 1 1 1 1 1 1 1 1 1## [1] 1901 1902 1903 1904 1905 1906 1907 1908 1909 1910# create dates using ISOdate function
(october1s <- ISOdate(year = october.year,
month = october.month,
day = october.day))## [1] "1901-10-01 12:00:00 GMT" "1902-10-01 12:00:00 GMT" "1903-10-01 12:00:00 GMT" "1904-10-01 12:00:00 GMT" "1905-10-01 12:00:00 GMT"
## [6] "1906-10-01 12:00:00 GMT" "1907-10-01 12:00:00 GMT" "1908-10-01 12:00:00 GMT" "1909-10-01 12:00:00 GMT" "1910-10-01 12:00:00 GMT"# use lubridate to calculate the julian date for October 1 for all years of interest;
(julians <- yday(october1s))## [1] 274 274 274 275 274 274 274 275 274 274Notice in that in typical years, the Julian date for October 1 is 274, while in leap years the Julian date is 275.
Our harvest data consists of records from multiple years, and we need to match each record’s harvest year with the proper October 1 Julian date. The key word there is match, and conveniently the match function can be used for this purpose. The easiest way to learn about the match function is to see it in action…
## [1] 1901 1901 1901 1901 1901 1901 1901 1901 1901 1901## [1] 1901 1902 1903 1904 1905 1906 1907 1908 1909 1910# find the year index for each harvest record; store as an object called indices
indices <- match(x = harvest$year, table = october.year)
# look at the first few records of indices so you can see what's going on
head(indices, n = 10)## [1] 1 1 1 1 1 1 1 1 1 1Notice that our first harvest record was harvested in 1901. The match function tells us that this is index 1 in the october.year vector. Our second harvest record was harvested in 1901. The match function tells us that this is index 1 in the october.year vector.
The match function is a very useful function, and one you may use frequently in your work. Make sure to read the helpfile. The function returns an index, or position, of something you are looking for. The first argument, x is what we’d like to match (the year of each harvest record). The second argument, table is a vector that we’d like to match against.
Now we can create a new column called first.day (opening day of the harvest season) and populate this column with the proper October 1 julian date for each harvest record, depending on what year the record was collected.
To see what happened, let’s randomly sample 10 harvested animals (rows) from our dataset:
# collect 10 random samples of rows from the harvest dataframe
samples <- sample(nrow(harvest), size = 10, replace = FALSE)
# look at the samples
harvest[samples, ]## hunter.id age sex individual species date town length weight method color fur month year julian first.day
## 11405 230 3 m 11405 Tauntaun 1909-10-18 GREENSBORO 390.71 865.9785 lightsaber gray long 10 1909 291 274
## 6229 99 1 m 6229 Tauntaun 1906-12-27 IRASBURG 323.32 726.3909 bow white medium 12 1906 361 274
## 4634 679 1 m 4634 Tauntaun 1905-11-03 BROWNINGTON 321.90 828.3958 muzzle gray long 11 1905 307 274
## 9385 622 2 f 9385 Tauntaun 1908-11-03 WARDSBORO 303.70 622.6445 bow white long 11 1908 308 275
## 6012 281 1 m 6012 Tauntaun 1906-11-30 BRIGHTON 467.89 1111.7601 lightsaber gray medium 11 1906 334 274
## 8986 583 1 m 8986 Tauntaun 1908-11-27 BROOKLINE 475.68 1054.8944 bow gray medium 11 1908 332 275
## 5304 489 3 m 5304 Tauntaun 1905-10-12 RYEGATE 449.11 1127.5824 bow white medium 10 1905 285 274
## 10634 674 1 m 10634 Tauntaun 1909-12-12 CHITTENDEN 367.67 916.6662 lightsaber gray medium 12 1909 346 274
## 12086 377 1 m 12086 Tauntaun 1910-11-22 ROCHESTER 432.76 1226.5248 bow gray short 11 1910 326 274
## 8890 611 1 m 8890 Tauntaun 1908-11-18 LANDGROVE 377.65 1033.6520 bow white short 11 1908 323 275The column “first.day” gives the julian date for October 1st in each year.
Finally, we can create a new column called day.of.season which provides the day of the harvest season in which each animal was harvested.
# calculate the day of season in which the animal was harvested
harvest$day.of.season <- harvest$julian - harvest$first.day + 1
# drop the column called first.day
harvest$first.day <- NULL
# look at the first few records
head(harvest)## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 1 285 1 f 1 Tauntaun 1901-10-08 STANNARD 277.10 699.3044 lightsaber white long 10 1901 281 8
## 2 56 1 f 2 Tauntaun 1901-11-13 BALTIMORE 311.86 718.1022 muzzle gray long 11 1901 317 44
## 3 549 1 f 3 Tauntaun 1901-12-26 HANCOCK 274.84 550.6420 bow white long 12 1901 360 87
## 4 296 1 f 4 Tauntaun 1901-12-11 ST. ALBANS TOWN 266.70 699.5951 muzzle gray long 12 1901 345 72
## 5 521 1 f 5 Tauntaun 1901-11-21 WHEELOCK 271.84 613.3350 bow gray long 11 1901 325 52
## 6 355 1 f 6 Tauntaun 1901-10-18 PEACHAM 301.03 851.4671 lightsaber grey short 10 1901 291 18So, our first record is a tauntaun harvested on day 8 of the harvest season. The calculation for this was the julian day of harvest minus the julian date for October 1 (first.day, or opening day). We added the number 1 to this result so that October 1 is Day 1 (and not Day 0).
There may be simpler ways to calculate the day.of.season column, but we wanted to introduce the match function to you.
It’s often very useful to plot a frequency histogram of the data. Let’s try that now with the hist function:
hist(x = harvest$day.of.season,
breaks = 10,
xlab = "Day of Season",
col = "deepskyblue1",
main = "Numbers of Tauntauns Harvested by Day of the Season")
This hist function is a quick way to generate histograms, and has several arguments that allow you to customize it to your liking. Here, we entered “Day of Season” for the x-label argument (xlab), set the bar color to “deepskyblue1”, and entered a title for the chart. If you want a different color, try the colors function (with no arguments) to see what options are available.
6.6.7 town = town in which the animal was harvested
Vermont has 255 towns, and your organization is required to summarize the harvest data by town as part of your annual report (we’ll do this in future chapters). We don’t need to see all 255 records now, but let’s look at the first 10 records. Since it’s a data frame, we need to use the proper indexing to extract information, which is [Rows, Columns].
## [1] STANNARD BALTIMORE HANCOCK ST. ALBANS TOWN WHEELOCK PEACHAM AVERYS GORE SUDBURY MOUNT HOLLY
## [10] BROOKFIELD
## 255 Levels: ADDISON ALBANY ALBURGH ANDOVER ARLINGTON ATHENS AVERILL AVERYS GORE BAKERSFIELD BALTIMORE BARNARD BARNET BARRE CITY BARRE TOWN ... WORCESTERWe can tell that R is storing the data in this column as a factor because 255 levels are identified. We’ll assume this is fine and that there are no typos so that we can move on.
6.6.8 length = length of harvested animal
Let’s look again at the column names for our object, harvest:
## [1] "hunter.id" "age" "sex" "individual" "species" "date" "town" "length" "weight"
## [10] "method" "color" "fur" "month" "year" "julian" "day.of.season"The next columns are length and weight. Let’s look at how the values are stored, beginning with length:
## num [1:13351] 277 312 275 267 272 ...You can see that R stores the data as numbers. We could use the summary function once again to inspect the data, but instead we’ll introduce you to the boxplot function.
Here, we will look at all of the data in our length vector.
## length
## Min. : 228.8
## 1st Qu.: 301.6
## Median : 343.4
## Mean : 355.1
## 3rd Qu.: 403.0
## Max. :4591.8Boxplots are oh-so-very helpful in looking at a column of numeric data. Do you recall how to interpret them? The graphic below is from a tutorial by Flowing Data, where they state:
The box-and-whisker plot is an exploratory graphic, created by John W. Tukey, used to show the distribution of a dataset (at a glance). Think of the type of data you might use a histogram with, and the box-and-whisker (or box plot, for short) could probably be useful.
Figure 6.4: Source: http://flowingdata.com.
Before we interpret this plot, imagine that we took all of our dead Tauntauns and ordered them shortest to tallest. This ordering is critical. Now look at the boxplot. The thick line in the middle of the plot is the median value…the median is the datapoint in the middle of the Tauntaun lineup: 50% of the data have values greater than the median, and 50% have values less than the median. Then we see the upper and lower quartiles in our boxplot. The word “quartiles” sounds like “quarters”. You’d be right in assuming that the upper quartile is the datapoint (in our ordered vector) where 25% of the data have a greater value, and 75% have a lower value. Same goes for the lower quartile, but in reverse. The tails of our boxplot give the maximum and minimum values, excluding the outliers. And the outliers themselves are identified as dots. Looks like we have a few! The outliers are identified as those records that are less than 3/2 times the lower quartile, or greater than 3/2 times the upper quartile.
Another great feature of the boxplot function is that you can store its outputs as a new object. Next, we’ll use the exact same code as we used earlier, but just assign it to the object box.length.out.

## $stats
## [,1]
## [1,] 228.750
## [2,] 301.620
## [3,] 343.370
## [4,] 403.005
## [5,] 545.700
##
## $n
## [1] 13351
##
## $conf
## [,1]
## [1,] 341.9836
## [2,] 344.7564
##
## $out
## [1] 2960.6 3599.2 4158.3 4591.8 4083.6 4135.3
##
## $group
## [1] 1 1 1 1 1 1
##
## $names
## [1] "length"This is useful because you can now access this object and retrieve information from it. Can you recognize this as a list? Let’s confirm our intuition:
## List of 6
## $ stats: num [1:5, 1] 229 302 343 403 546
## $ n : num 13351
## $ conf : num [1:2, 1] 342 345
## $ out : num [1:6] 2961 3599 4158 4592 4084 ...
## $ group: num [1:6] 1 1 1 1 1 1
## $ names: chr "length"The outliers are stored in a vector within the list called out. The stats portion of this list is a matrix, which is storing some of the information you obtained with the summary function. If you want to extract the stats (or any other) element from this list, you could use one of the different extractor methods to do so:
## [,1]
## [1,] 228.750
## [2,] 301.620
## [3,] 343.370
## [4,] 403.005
## [5,] 545.700As the Tauntaun expert, you know that males and females differ in size, and are concerned about any lengths greater than or equal to 549 units for males and 374 for females. How can we look at the data by sex?
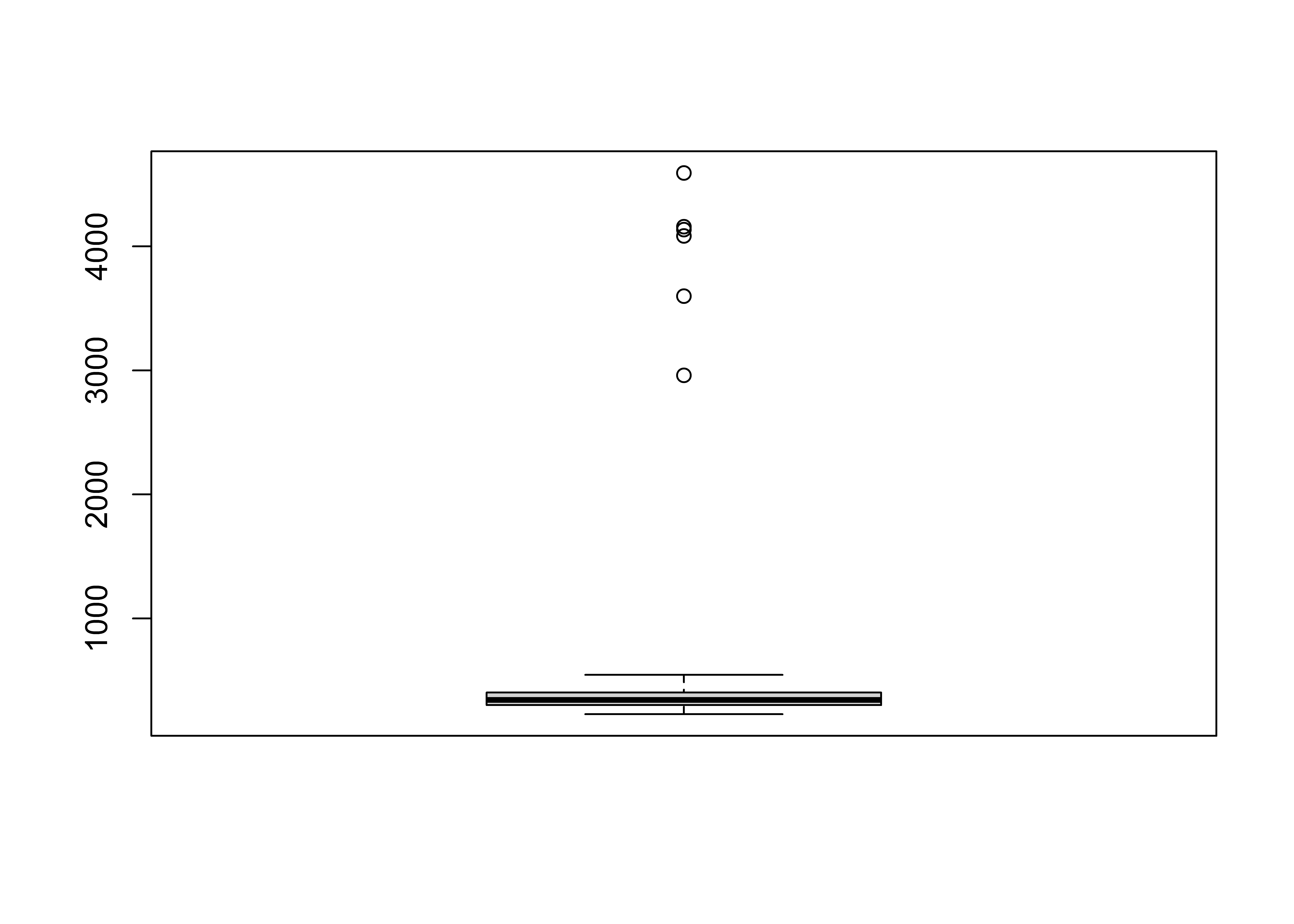
The boxplot function can be handy for looking at how the continuous data (lengths) are distributed among factors of another column. For example, if we want to create a boxplot of length for each factor of sex (“m” and “f”), we could use R’s formula notation:
This is the first time we’ve seen R’s formula notation, so let’s talk about it for a moment. You’ve seen equations that set y as a function of x, right? In R, that is written y ~ x, where y is the dependent variable and x is the predictor or independent variable. For example, if you want to predict weight as a function of length, you would write weight ~ length. So ~ (known as a ‘tilde’) means “as a function of” or “is predicted by”.
Here, we use the boxplot function and ask R to give us boxplots of length as a function of sex….we end up with a boxplot for each sex individually. The boxplot function has several other arguments that can be used to add axes, titles, etc. to your plot . . . we’ll revisit this in future chapters. For now, we can see that outliers occur for both sexes. Now what? Are they typos? Or are they just really, really long Tauntauns?
The first thing to do is to extract the suspicious records and inspect them. A great function to carry out this task is the which function – this function requires that you “ask a question” that can be answered TRUE or FALSE. R will return the indices for the TRUE responses. We need to treat males and females differently because different “rules” apply as to what constitutes an outlier (i.e. greater than or equal to 549 for males versus 374 for females). First, let’s deal with the male outliers:
# find the rows with lengths greater than or equal to (>=) 549 AND sex = "m"
which(harvest['length'] >= 549 & harvest['sex'] == "m")## [1] 4493 4513 7683 7985 13308What does R return? These are indices, or positions, within the length vector that meet both conditions: the length is greater than or equal to 549 AND the sex is equal to ‘m’. Let’s store these indices as a new object called m.outliers (short for male outliers). Then, let’s extract these records from our harvest dataframe (which consists of rows and columns) using the [R,C] extraction method, pulling out only those rows which contain the outliers:
# assign these indices to the vector, m.outliers
m.outliers <- which(harvest['length'] >= 549 & harvest['sex'] == "m")
# extract the rows identified, and look at all columns associated with these rows
harvest[m.outliers,]## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 4493 255 1 m 4493 Tauntaun 1905-10-22 TOPSHAM 3599.2 992.6929 muzzle gray short 10 1905 295 22
## 4513 533 1 m 4513 Tauntaun 1905-10-04 PROCTOR 4158.3 1148.9063 bow gray medium 10 1905 277 4
## 7683 596 1 m 7683 Tauntaun 1907-10-05 PERU 4591.8 1196.7814 muzzle gray medium 10 1907 278 5
## 7985 566 2 m 7985 Tauntaun 1907-10-27 BROOKLINE 4083.6 1109.0055 bow gray medium 10 1907 300 27
## 13308 71 5 m 13308 Tauntaun 1910-12-08 SOUTH HERO 4135.3 871.0144 muzzle gray short 12 1910 342 69There is nothing obviously wrong with these rows other than the erroneous length values…they are much too large to be actual Tauntauns, and there is no apparent “fix” to the data. We could either delete the entire record for each, or delete just the lengths. Let’s just delete the lengths so we can still use the rest of the data.
Data frames cannot hold NULL values (blank entries with absolutely nothing in them), so the next best would be to replace the erroneous lengths with NA values, which stands for Not Available. We reassign these values by extracting the correct rows (identified by m.outliers and column (length), and then setting the value to NA.
# those lengths are all greater than 549, so now we assign NA values
harvest[m.outliers,'length'] <- NAFor female Tauntauns, we are suspect of any lengths > 374 units. So we’ll repeat our exercise to replace the flawed records with NA:
# find the rows with lengths greater than or equal to 374 AND sex = "f"
which(harvest['length'] >= 374 & harvest['sex'] == "f")## [1] 3191# assign these indices to the vector, f.outliers
f.outliers <- which(harvest['length'] >= 374 & harvest['sex'] == "f")
# extract the rows identified, and look at all columns
harvest[f.outliers,]## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 3191 31 1 f 3191 Tauntaun 1904-10-11 BROOKFIELD 2960.6 653.9171 bow gray long 10 1904 285 11# select the rows f.outliers, and the column **length**
harvest[f.outliers,'length'] <- NA
# look at the resulting length data
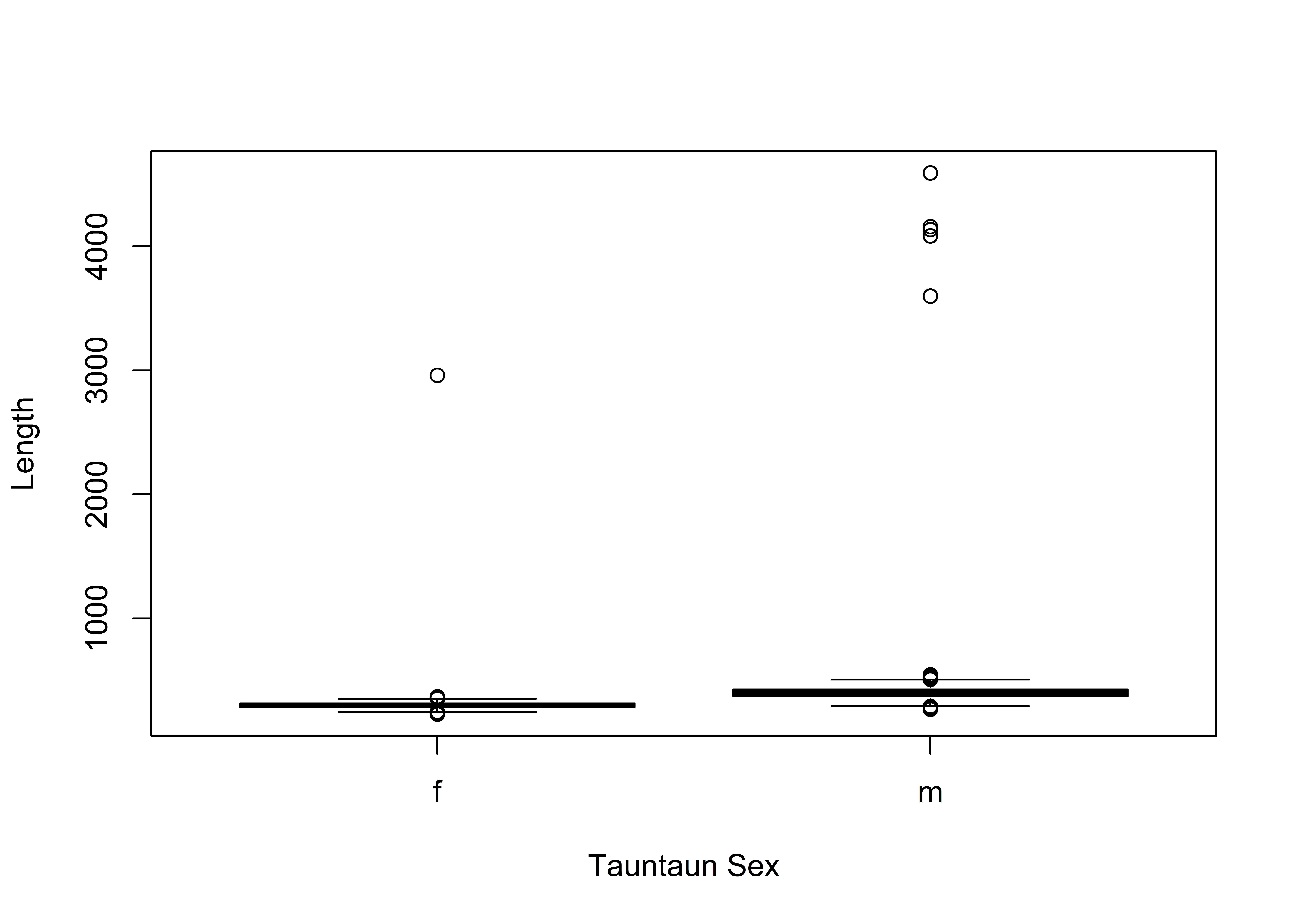
boxplot(harvest[,'length'] ~ harvest[,'sex'])
That’s more like it. Of course, the challenge with outliers is deciding how to handle them. Here, they were obviously typos. But what if a Tauntuan is just borderline too big for your comfort? The decision on how to handle this kind of outlier is not so clear cut…it might be a good time to chat with your friendly neighborhood statistician, or maybe a colleague who might shed some light on why Tauntauns are larger than expected. As long as they are not typos or measurement errors, outliers may be very interesting observations reflecting natural variation!
6.6.9 weight = weight of harvested animal (arbitrary units)
Now let’s turn our attention to the column weight, which also stores numbers. We’ll use the boxplot function again here to spot potential outliers. Here, we expect that weights for males and females should differ, and wish to use the boxplot function for each sex separately.
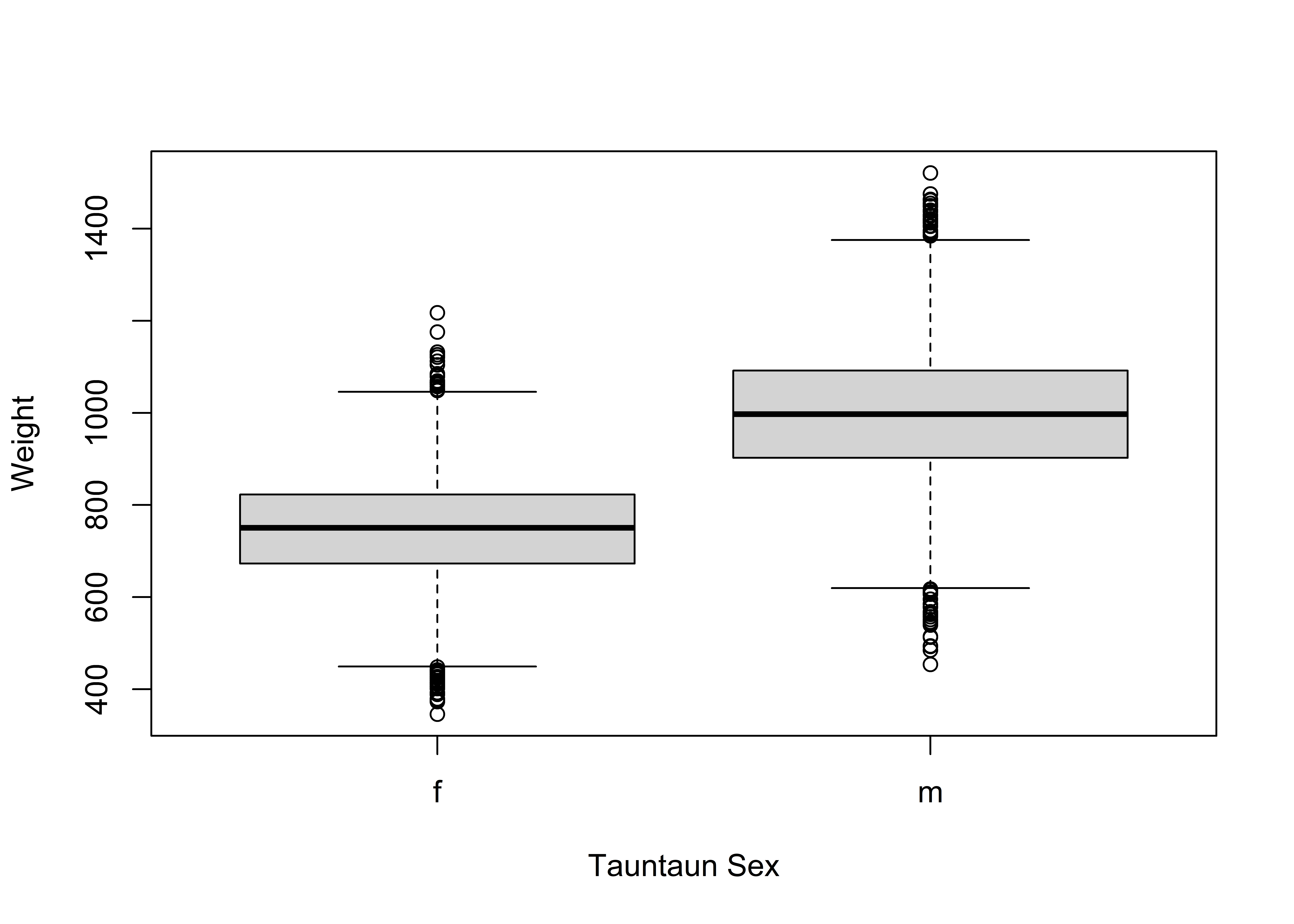
Although there are a few outliers, you have no reason to expect that these may be errors. None of these records appear to be “flags” to you, but it would be nice to see the data in a histogram for each sex.
We could subset the data by sex and then use the hist function as we have previously. But here we’ll introduce you to a graphics package called ggplot2, which is very useful for creating graphics of all kinds and is another tidyverse package (authored by Hadley Wickham and colleagues).
To be sure, there are many ways to plot data in R. The base R program’s plotting functions are sufficient for most cases, but here we will use ggplot2 because it has great documentation that can propel you much further than this book. In a nutshell, a the ggplot approach builds a graph piecewise. The main function, ggplot requires a dataframe or tibble for the first argument. The second argument provides the mapping of your graph: what goes on the axes. From there, you add the maps layers, such as a histogram. A faceting specification describes how to break up the data into subsets, such as producing a histogram by Tauntaun sex. Here is our sample code:
# load ggplot2
library(ggplot2)
ggplot(data = harvest, aes(x = weight, fill = sex)) +
geom_histogram(binwidth = 20) +
facet_wrap(~sex)
There are many, many ggplot options, and if this is your graphing package of choice, we encourage you to work through the free ggplot2 book.
The data in this column look fine to you as the Tauntaun biologist, so let’s move along.
6.6.10 method = method of harvest
Our next column is called method, which gives the harvesting implement used by the hunter to kill a Tauntaun.
## 'data.frame': 13351 obs. of 1 variable:
## $ method: Factor w/ 3 levels "bow","lightsaber",..: 2 3 1 3 1 2 2 1 2 2 ...## [1] lightsaber muzzle bow <NA>
## Levels: bow lightsaber muzzleR is storing this column as a factor with 3 levels: “bow”, “muzzle” (for muzzleloader), and “lightsaber”. But what else is there? Do you see <NA>?
6.7 A Brief Interruption to Discuss NA and NULL
NA and NULL mean two different things. NA means that the data actually contains the value NA stored as a data entry. NULL means that the cell is empty. When you read in the harvest csv file, R filled in the NULL entries with NA’s because data frames cannot have NULLS. Importantly, the NA that R filled in is special type of value that cannot be used to index and subset the dataframe in the same way we have previously shown you.
Here’s an example to show you how this will not work as you might have expected. To find NA values in our dataframe, we do NOT index based on a logical operator like this:
# we mistakenly hope this will return the records that have NA's
NAsubset <- harvest['method'] == NA
# look at only the first five records, but notice that all of the records are set to NA!
head(NAsubset)## method
## [1,] NA
## [2,] NA
## [3,] NA
## [4,] NA
## [5,] NA
## [6,] NAInstead, we have to use a special function that searches for NA values, is.na:
# this returns a vector filled with TRUE or FALSE, depending on whether the value is NA
na.logical <- is.na(harvest['method'])
head(na.logical)## method
## [1,] FALSE
## [2,] FALSE
## [3,] FALSE
## [4,] FALSE
## [5,] FALSE
## [6,] FALSEThe is.na function returns a vector filled with TRUE or FALSE for each and every record, depending on whether the value is NA. We can use the true false vector to view the associated records within harvest (select rows where method is NA, and all columns). Notice we’re nesting it in the head() function to keep output to a minimum:
## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 103 412 1 f 103 Tauntaun 1901-10-11 SHAFTSBURY 320.32 747.7223 <NA> white long 10 1901 284 11
## 174 516 1 f 174 Tauntaun 1901-12-05 ROCHESTER 289.28 731.2382 <NA> gray long 12 1901 339 66
## 219 687 1 f 219 Tauntaun 1901-11-29 BARNARD 308.23 1057.4002 <NA> gray short 11 1901 333 60
## 221 571 1 f 221 Tauntaun 1901-11-21 AVERYS GORE 317.43 861.7146 <NA> gray short 11 1901 325 52
## 223 609 1 f 223 Tauntaun 1901-10-05 SHOREHAM 365.51 780.5430 <NA> gray medium 10 1901 278 5
## 251 531 1 f 251 Tauntaun 1901-11-09 WOODBURY 304.55 748.0314 <NA> white short 11 1901 313 40Another way to subset your data to snoop out the NA values is to use the subset function, another very useful function that you will use frequently. Take a look at the helpfile, and recognize that a few key arguments are needed: x is the object to be subsetted, subset is a logical expression that evaluates to TRUE or FALSE for each and every element. There are other arguments too, but for our purposes we can get by just specifying these two arguments.
## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 103 412 1 f 103 Tauntaun 1901-10-11 SHAFTSBURY 320.32 747.7223 <NA> white long 10 1901 284 11
## 174 516 1 f 174 Tauntaun 1901-12-05 ROCHESTER 289.28 731.2382 <NA> gray long 12 1901 339 66
## 219 687 1 f 219 Tauntaun 1901-11-29 BARNARD 308.23 1057.4002 <NA> gray short 11 1901 333 60
## 221 571 1 f 221 Tauntaun 1901-11-21 AVERYS GORE 317.43 861.7146 <NA> gray short 11 1901 325 52
## 223 609 1 f 223 Tauntaun 1901-10-05 SHOREHAM 365.51 780.5430 <NA> gray medium 10 1901 278 5
## 251 531 1 f 251 Tauntaun 1901-11-09 WOODBURY 304.55 748.0314 <NA> white short 11 1901 313 40You should see that the same result is returned. A similar function exists to locate NULL values, is.null. But since NULL is not permitted in a data frame we don’t need to use it.
As an aside, have a look at the row names returned from the subset function. These are the original row numbers from the original harvest dataset. If you’d like to reset them, simply set the rownames to NULL, like this:
# subset the data again
harvest.subset <- subset(x = harvest, subset = is.na(method))
# look at the first few records, note the rownames
head(harvest.subset)## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 103 412 1 f 103 Tauntaun 1901-10-11 SHAFTSBURY 320.32 747.7223 <NA> white long 10 1901 284 11
## 174 516 1 f 174 Tauntaun 1901-12-05 ROCHESTER 289.28 731.2382 <NA> gray long 12 1901 339 66
## 219 687 1 f 219 Tauntaun 1901-11-29 BARNARD 308.23 1057.4002 <NA> gray short 11 1901 333 60
## 221 571 1 f 221 Tauntaun 1901-11-21 AVERYS GORE 317.43 861.7146 <NA> gray short 11 1901 325 52
## 223 609 1 f 223 Tauntaun 1901-10-05 SHOREHAM 365.51 780.5430 <NA> gray medium 10 1901 278 5
## 251 531 1 f 251 Tauntaun 1901-11-09 WOODBURY 304.55 748.0314 <NA> white short 11 1901 313 40# reset the rownames
rownames(harvest.subset) <- NULL
# look at the first few records, note the rownames
head(harvest.subset)## hunter.id age sex individual species date town length weight method color fur month year julian day.of.season
## 1 412 1 f 103 Tauntaun 1901-10-11 SHAFTSBURY 320.32 747.7223 <NA> white long 10 1901 284 11
## 2 516 1 f 174 Tauntaun 1901-12-05 ROCHESTER 289.28 731.2382 <NA> gray long 12 1901 339 66
## 3 687 1 f 219 Tauntaun 1901-11-29 BARNARD 308.23 1057.4002 <NA> gray short 11 1901 333 60
## 4 571 1 f 221 Tauntaun 1901-11-21 AVERYS GORE 317.43 861.7146 <NA> gray short 11 1901 325 52
## 5 609 1 f 223 Tauntaun 1901-10-05 SHOREHAM 365.51 780.5430 <NA> gray medium 10 1901 278 5
## 6 531 1 f 251 Tauntaun 1901-11-09 WOODBURY 304.55 748.0314 <NA> white short 11 1901 313 40Another shortcut to strip all records containing NA values is na.omit.
# create a dataframe call test that strips all records that contain NAs
test <- na.omit(harvest)
# look at its dimensions (rows, columns)
dim(test)## [1] 12833 16## [1] 13351 16This will omit all records that contain any NA values; sometimes this is what you want to do, but other times it is throwing out the baby with the bathwater (i.e., removing entire rows even when just a single column contains an NA). Alternatively, R has methods for ignoring NA, for example in creating summary statistics of a set of values.
Remember that R does not have an ‘undo’ button that can undo executed code! If you are unsure about what your code might produce, you should store the results as an object called “test” (or similar), run the code and then verify what is going on. If your code produces the intended results, then you can execute it on your real data.
6.7.1 color = fur color of the reptomammalian Tauntaun
Almost there . . . Almost there . . . The next column is called color. Let’s take a look:
## Factor w/ 3 levels "gray","grey",..: 3 1 3 1 1 2 3 1 3 3 ...## [1] white gray grey
## Levels: gray grey whiteThe ‘color’ column reports white, and two spellings for gray (gray and grey); we need to clean that up and use the “gray” version (with apologies to any British readers). Let’s use the table function to quickly summarize the numbers:
##
## gray grey white
## 8193 1306 3852It looks like some of your data entry volunteers went with “grey”, while others went with “gray”. Typically, this fix would be done in a single line as we did earlier with the species column.
To fix these, however, we’ll display the components separately and assemble them into a single line afterward to sharpen our coding skills. First, we’ll extract the indices of the rows, and then reassign the values. The key will be that we use logical operators to identify which rows meet our criteria, and from that we will create a logical matrix for indexing.
# use the 'which' function and "equal to" operator to find row indices of "grey" Tautauns
change <- which(harvest$color == 'grey')
# look at a few entries in change, note that they are indices within the color column
change[1:10]## [1] 6 20 21 26 28 44 50 51 59 63# replace the "greys" with "grays", using the R and C indexing
harvest[change,'color'] <- 'gray'
# check again
table(harvest$color)##
## gray grey white
## 9499 0 3852As a double check, we can determine how many rows should be “gray” by counting those records that are NOT equal to white. To do this, we use a “not equal to” operator, which is ‘exclamation point equal sign’, like this:
is.Gray <- harvest['color'] != 'white'
# look at the first few records of the dataset
head(harvest$color, n = 5)## [1] white gray white gray gray
## Levels: gray grey white## [1] FALSE TRUE FALSE TRUE TRUE## [1] 9499In this code, we first evaluate whether every single element in the object meets our logical criteria (not white). Then we sum the logical values, where TRUE = 1 and FALSE = 0.
Note that we could have done all of this in one step!
# see if you can identify what is going on with this code
harvest[harvest['color'] == 'grey', 'color'] <- 'gray'This is just Row and Column indexing in action [r,c]. We are subsetting the harvest dataframe by rows and columns. Which rows? All rows where the column color == gray. Which columns? The column called color. In the resulting subset, we set the data to gray.
We are treating the process of subsetting lightly, but it is perhaps the single most critical skill to master in R, as it allows you to precisely manipulate your data without introducing error. You should practice selecting a single fur color at a time or a single age class until you understand the syntax you must use. Once mastered, this syntax can be expanded to include a wide variety of expressions.
Now let’s check the unique values of the colors column, and it should report no more ‘grey’ values.
## [1] white gray
## Levels: gray grey whiteNotice we don’t have any more ‘grey’ values, but the factor level ‘grey’ is still being stored.
Let’s confirm this by peeking at the raw data again:
## [1] white gray white gray gray gray white gray white white
## Levels: gray grey whiteAlthough in this instance you can ignore this, there are times when you need the factors to exactly represent the data (such as if you want to build a contingency table with table). We can drop the unused level by recasting the factor levels from the current values using the factor function, and we can look at the factor levels using the levels function.
First, read the levels helpfile, then read the factor helpfile. There, we learn that “The function factor is used to encode a vector as a factor (the terms ‘category’ and ‘enumerated type’ are also used for factors). If argument ordered is TRUE, the factor levels are assumed to be ordered.” When you use the factor function, you tell R what levels are currently represented in your dataset, and you also can give them labels. If the levels differ from the labels, the labels will replace the existing levels.
## [1] "gray" "grey" "white"# use the factor function to reset the factors
harvest$color <- factor(harvest$color,
levels = c("white", "gray"),
labels = c("White", "Gray"),
ordered = FALSE)
# look at the structure again
str(harvest$color) ## Factor w/ 2 levels "White","Gray": 1 2 1 2 2 2 1 2 1 1 ...## [1] White Gray White Gray Gray Gray White Gray White White
## Levels: White GrayThe labels can be used by graphics functions. For example, if we use the ggplot function to create a histogram of tauntaun color morphs, you’ll see the labels (which we capitalized simply for appearance) in use:
ggplot(data = harvest, aes(x = color)) +
geom_bar(stat = "count", fill = "steelblue") +
xlab("Color morph") +
ylab("Total count") 
6.7.2 fur: Tauntaun fur length
Finally…our last column. We’re out of breath, so let’s dive right in.
## Factor w/ 3 levels "long","medium",..: 1 1 1 1 1 3 3 1 2 1 ...##
## long medium short
## 4604 4334 4413Here we see another factored column, also with three factors. We can also see that these factors should be ordered from shortest to longest, i.e. short, medium, and long. That way, any of the plots we create will show the responses in this order, for example from left to right along the x-axis. Again, the factor function can provide some help, this time with the “order” argument.
## [1] "long" "medium" "short"# use the factor function to reset the factors
harvest['fur'] <- factor(harvest[,'fur'],
levels = c("short", "medium", "long"),
labels = c("Short", "Medium", "Long"),
ordered = TRUE)
# look at the structure again
str(harvest[,'fur']) ## Ord.factor w/ 3 levels "Short"<"Medium"<..: 3 3 3 3 3 1 1 3 2 3 ...##
## Short Medium Long
## 4413 4334 46046.8 Stepping through Rows and Columns with Apply Functions
We’ve stepped through each column of our harvest dataset one by one, and as tedious as that may seem, it’s the best way to really understand what’s in your dataset and to be sure that it is free of errors. This process is known as “data wrangling”. Wikipedia tells us that “Data wrangling, sometimes referred to as data munging, is the process of transforming and mapping data from one”raw" data form into another format with the intent of making it more appropriate and valuable for a variety of downstream purposes such as analytics."
We’ve seen that R has a group of functions known as “apply functions” that repeat an action on all rows or columns of a table, on all elements of a vector, or on all elements of a list. We touched on the apply functions previously. Let’s use the sapply function to quickly double-check the data type in each column.
# use the sapply function to walk through each column of harvest and return the class
sapply(X = harvest, FUN = class)## $hunter.id
## [1] "integer"
##
## $age
## [1] "integer"
##
## $sex
## [1] "factor"
##
## $individual
## [1] "integer"
##
## $species
## [1] "character"
##
## $date
## [1] "Date"
##
## $town
## [1] "factor"
##
## $length
## [1] "numeric"
##
## $weight
## [1] "numeric"
##
## $method
## [1] "factor"
##
## $color
## [1] "factor"
##
## $fur
## [1] "ordered" "factor"
##
## $month
## [1] "numeric"
##
## $year
## [1] "numeric"
##
## $julian
## [1] "numeric"
##
## $day.of.season
## [1] "numeric"You can see that the sapply function returned the class for each and every column of the harvest dataframe. The arguments of sapply are “X”, the object to work on (in our case, harvest), and “FUN”, the function that you want to run on each element within harvest (in our case, class). Don’t forget that harvest is a data frame, and therefore a list of vectors. So sapply will work through each vector, and execute the function you specify. It will then attempt to simplify the result from a list to either a vector or a matrix.
You may recall there are other “apply” functions too, such as lapply and apply, tapply (see Chapter 5.9).
That wraps up our “walk” through the harvest dataset. In practice, you will find a rhythm of checking the data types, looking for errors, fixing the errors, and confirming the results. Get used to this! A large proportion of any data scientists job involves data wrangling.
6.9 Save the Cleaned Data as harvest_clean.RData
We’ve reviewed each column in our harvest csv files, and we made several changes to the harvest dataset. The last thing to do is to write the perfectly cleaned file to your datasets folder. That way, it will always be cleaned and ready for any future analyses.
We could save it as another csv file with write.csv, but can you guess why this might not be wise? You got it….when we read it in again with read.csv, we’ll have to re-run our code to set the correct data types for each column. Why go to that trouble? Let’s instead save it as an RData file with the save function:
# create a R object called harvest_clean.RData)
save(harvest, file = 'datasets/harvest_clean.RData')
# verify that the file exists
file.exists('datasets/harvest_clean.RData')## [1] TRUEThis saves the object in our working environment called harvest as an .RData file. (Another file format is Rda, which is a short name for RData.) Look for this new file in your Files tab. You should see a file called harvest_clean.RData. This file stores the current state of the object called harvest at the time you save it. By storing the final, cleaned version of harvest, you can retain the original data as a csv file, the script that you used to clean it, and the final output file that is ready for use in the next chapter. You’ll see that you can load this .RData file back into your global environment, the object harvest will appear in the exact same state as when you saved it.
Don’t forget to save your script file! This will be a record of all of the changes you have made to the original data. It might come in handy next year when you have to clean new records.
6.10 Read in the Hunter CSV file, and Save as hunter_clean.RData
Our harvest data frame has the hunter ID in our harvest data, but no other information about them (such as their age, sex, or where they live). Remember that second csv file you inherited? It contains records about the hunters themselves. Let’s read that into R, and take a look at this dataset.
# read in the Hunter table and check for errors
hunter <- read.csv(file = "datasets/hunter.csv", stringsAsFactors = TRUE)
# look at the structure
str(hunter)## 'data.frame': 700 obs. of 3 variables:
## $ hunter.id : int 1 2 3 4 5 6 7 8 9 10 ...
## $ sex.hunter: Factor w/ 2 levels "f","m": 2 2 2 2 2 1 2 2 2 2 ...
## $ resident : logi TRUE FALSE TRUE TRUE FALSE TRUE ...## hunter.id sex.hunter resident
## Min. : 1.0 f: 94 Mode :logical
## 1st Qu.:175.8 m:606 FALSE:138
## Median :350.5 TRUE :562
## Mean :350.5
## 3rd Qu.:525.2
## Max. :700.0There isn’t much to fix here, the columns hold simple information: the hunter’s id, the sex of the hunter, and a column called resident which indicates whether the hunter is a resident of Vermont or not. Nothing looks unexpected in the summary or structure. Let’s take a look at the first 6 rows of data:
## hunter.id sex.hunter resident
## 1 1 m TRUE
## 2 2 m FALSE
## 3 3 m TRUE
## 4 4 m TRUE
## 5 5 m FALSE
## 6 6 f TRUEThis gives you a good feeling for this dataset, and no changes are necessary. But, to keep things consistent, let’s create an .RData file for the hunters as well. That way, in the next chapter, we can load the two .RData files (which are R objects that are cleaned and error free), and begin working on them directly.
# create a R object called hunter_clean.RData)
save(hunter,file = 'datasets/hunter_clean.RData')
# check to see if the file exists
file.exists("datasets/hunter_clean.RData")## [1] TRUE6.11 Next Steps…
Terrific job! We’ve learned several new functions in this chapter, and had lots of opportunities to practice indexing. Working with data will likely occupy a lot of your time as the tauntaun biologist, and we’ve just scratched the surface. For a more comprehensive treatment of data cleaning, check out this article on the Cran website.
In the next chapter, we’ll learn how to merge our two files together, and then also add in some spatial data to boot. We’ll practice plotting, sorting, and subsetting data in new ways as well. Then we will save the final dataset as an .RDS file, which will store our final dataset (cleaned and merged, ready for some serious analysis). See you there.