Systemic Acquired Resistance (SAR) in Plants
The oomycete
Hyaloperonospora parasitica invading wild-type Arabidopsis
thaliana leaf tissue (formerly called Peronospora parasitica). |
Plants have both constitutive and induced mechanisms that aid them in defense against disease. Following a localized infection, many plants have been shown to undergo a systemic reaction that leads to broad-spectrum resistance to a diverse variety of pathogens, including fungi, bacteria and viruses. Because the entire plant manifests the resistance response, this form of induced resistance is called systemic acquired resistance or SAR. Interestingly, the same response can be triggered by exposure to any of a diverse variety of pathogens, suggesting that multiple sensory mechanisms converge upon a common response pathway. The onset of SAR has been shown to be accompanied by the accumulation of salicylic acid, a wide variety of mRNA species and their encoded protein products (for review see Delaney, 2004).
An H.
parasitica sporangiophore (asexual fruiting body). |
Salicylic acid (SA) levels increase in plants undergoing SAR. Further, application of SA induces the same set of genes and spectrum of resistance specificities as found in biologically-induced SAR. These observations led to the hypothesis that SA acts as a signal that triggers SAR. This model was validated by experiments in which tobacco plants were engineered to express a bacterial salicylate hydroxylase gene, whose product degrades SA. Plants expressing this gene were unable to accumulate SA, and importantly were unable to mount an SAR response, demonstrating that SA accumulation was required for induction of SAR (Gaffney et al, 1993, Science 261, 754).
Other experiments were conducted using transgenic Arabidopsis thaliana plants expressing salicylate hydroxylase. Like the tobacco plants described above, these plants were unable to induce SAR genes to high levels. Remarkably, we found that NahG plants were also defective in a distinct form of disease resistance, namely, race-specific or genetically-determined resistance (more details). Like the tobacco plants described above, transgenic Arabidopsis thaliana plants expressing salicylate hydroxylase were compromised in the induction of SAR genes. However, when examined carefully, these plants were also shown to permit growth of normally incompatible races of both fungal and bacterial pathogens, indicating that SA also plays a role in mediating genetically- determined resistance. Together, these results indicate that a common SA-dependent pathway exists that controls both SAR and gene-for-gene resistance (Delaney et al, 1994). Our later work showed also that in some cases, the NIM1/NPR1 gene described below, is also needed for full expression of gene-for-gene resistance (Rairdan et al., 2002).
H. parasitica
mycelia depositing haustoria
within Arabidopsis cells. |
To facilitate genetic dissection of the pathways controlling induced resistance to disease, we explored the SAR response in Arabidopsis thaliana, with the aim of using this plant in genetic experiments. Arabidopsis is attractive due to the wealth of genetic, molecular and pathological resources available. Other useful tools were developed by the discovery of synthetic chemicals that mimic the action of salicylic acid, in that they induce the same set of SAR-related genes and lead to the same spectrum of resistance as found in biologically-induced SAR. One such chemical is called INA, and was used in an effective mutant screen to identify mutations that disabled the pathway controlling SAR genes and resistance. This screen led to the discovery of the non-inducible immunity (nim) mutant class. Several plant lines were identified with mutations in the NIM1 gene. Plants with mutations in this gene retain the ability to accumulate SA in response to pathogen, yet no longer induce SAR genes or resistance, suggesting that the NIM1 gene product acts as a regulatory component in the signal transduction pathway controlling SAR(Delaney et al, 1995). In collaboration with colleagues at Novartis Inc., the NIM1 gene was cloned, and found to encode a protein that shows homology to the I-kappa-B family of transcriptional factor repressors (Ryals et al, 1997). I-KB proteins function in regulating gene expression in organisms as diverse as mammals and fruit flies, and their presence in plants suggests an ancient and conserved mechanism of gene regulation. Other labs independently identified the gene (named NPR1 by X. Dong, Duke University, or SAI1 by D. Klessig, then at Rutgers), and Dong's group also independently clone the gene (Cao et al., 1997) (References listed within Delaney, 1994).
A mutant screen developed in John Ryals' lab at Ciba Plant Biotechnology, set out to identify mutant plants with a phenotypes of de-regulation of SAR genes. We noted that such mutants often had an additional phenotype of necrotic lesions on their leaves. The presence of lesions and expression of SAR genes in these mutants is reminiscent of plants that had been 'triggered' to express SAR due to exposure to a necrosis-generating pathogen. At that time, Drs. J. Dangl and S. Grant (then at Max Delbruck Labs, Cologne) had identified a class of mutants based on the presence of necrotic lesions on their leaves. Based on the potential similarity of both collections of mutants, both labs arranged a collaborative investigation of the mutants. The question we asked was whether these mutants were undergoing an authentic disease-response reaction or whether they just superficially resembled diseased plants. Based on a variety of criteria, we concluded that five of the mutants indeed did act as though they perceived pathogen in its absence, which led to lesions, expression of SAR genes and resistance to pathogens. Based on these findings, such mutants were named lsd mutants, for their lesions-simulating-disease phenotype (Dietrich et al, 1994).
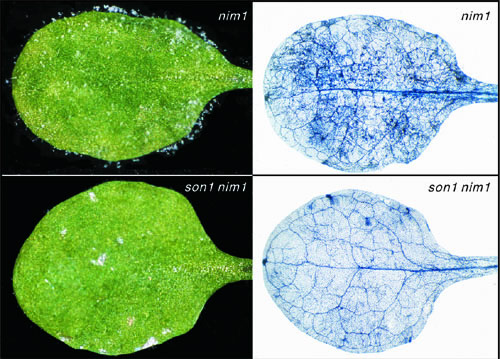 |
The son1 mutation confers SAR-independent resistance. Plants were inoculated with a virulent isolate of H. parasitica |
Mutant analysis indicates that SAR is just one form of induced resistance, and that others play important roles for maintaining plant health. To genetically identify mechanisms that underlie SAR-independent resistance (SIR), we have performed a number of mutant screens. One of these efforts involved a suppressor mutant screen to find mutations that re-activate disease resistance in normally defense-compromised nim1-1 plants. This work led to recovery of a mutation called son1 (suppressor of nim1-1) that was found to provide strong resistance to multiple pathogens independent of the NIM1/NPR1 pathway (Kim and Delaney, 2002a). Because resistance in son1 nim1 plants was not associated with any known markers for defense pathways, and does not require SA or NIM1/NPR1, is appears to be a novel form of disease resistance. We cloned the SON1 gene and found that it encodes an F-box protein, which are implicated in selecting proteins that will become ubititinated, and then proteolyzed (Kim and Delaney, 2002a). We postulate that SON1 targets for destruction a positive regulatior of SIR. We are pursuing our work with SON1 to establish whether it is a component of an SCF complex, which we predict based on the role of other better understood F-box proteins. We also are exploring the transcriptome of son1 and son1 nim1 plants to identify molecules that may be reponsible for the disease resistance phenotype observed in son1 plants.
The long-term goal of work in my lab is to elucidate mechanisms that plants use in regulating induced resistance to disease. This work in centered around three principal areas:
The most current publication list, with Citation Data can be viewed on Google Scholar: http://scholar.google.com/citations?user=XyUnP-8AAAAJ
Delaney, T.P. (2011) Salicylic Acid. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! 3rd Edition. Kluwer Academic Publishers, Dordrecht pp. 635-653.
Delaney, T.P., St.-Pierre, B., Li, Z. and Argueso, C., (2006) Identification and analysis of multi-layered disease resistance pathways in Arabidopsis. In Biology of Plant-Microbe Interactions, Vol. 5. F. Sánchez, C. Quinto, I.M. López-Lara, and O. Geiger (eds.) Intl. Soc. for Molec. Plant-Microbe Interactions. Merìda, Mexico, pp. 247-253. IS-MPMI Link
Delaney, T.P., Argueso, C., Kim H. S. and Ko, J.-H. (2004). Identification of disease resistance in Arabidopsis independent of systemic acquired resistance. In Biology of Plant-Microbe Interactions, Vol. 4. I. Tikhonovich, B. Lugtenberg and N. Provorov (eds.) Intl. Soc. for Molec. Plant-Microbe Interactions. St.-Petersburg, Russia, pp. 196-198. IS-MPMI Link
Buell, C. R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I. T., Gwinn, M. L., Dodson, R. J., Deboy, R. T., Durkin, A. S., Kolonay, J. F., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100, 10181-10186.Delaney, T.P. (2004) Salicylic Acid. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht pp. 635-653. Publisher Website
Peng J-L, Dong H-S, Dong H-P, Delaney TP, Bonasera JM, Beer SV (2003) Harpin-elicited hypersensitive cell death and pathogen resistance require the NDR1 and EDS1 genes. Physiol Molec Plant Pathol 62: 317-326.
Kim, H.S. and Delaney, T.P. (2002a) Arabidopsis SON1 regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14(7): 1469-82.
Kim, H.S. and Delaney, T.P. (2002b) Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant Journal 32: 151-163.
Rairdan, G.J. and Delaney, T.P. (2002) Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161(2): 803-11.
Rairdan, G.J., Donofrio, N.M. and Delaney, T.P. (2001) Salicylic acid and NIM1/NPR1-independent gene induction by incompatible Peronospora parasitica in Arabidopsis Molec. Plant-Microbe Interact. 14: 1235-46.
Donofrio, N.M. and Delaney, T.P. (2001) Abnormal callose response phenotype and hypersusceptibility to Peronospora parasitica in defense-compromised Arabidopsis nim1-1 and salicylate hydroxylase plants Molec. Plant-Microbe Interact. 14: 439-50. (journal cover)
Delaney, T. P., 2000. New mutants provide clues into regulation of systemic acquired resistance. Trends in Plant Science 5:2:49-51.
Dong, H., Delaney, T.P., Bauer, D.W. and Beer, S.V. 1999. Harpin induces systemic acquired resistance in Arabidopsis through the salicylic acid and NIM1-mediated signal transduction pathway. Plant J. 20: 207-15.
Delaney, T. P., 1997. Genetic dissection of acquired resistance to disease. Plant Physiol. 113: 5-12.
Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H-Y., Johnson, J., Delaney, T. P. Jesse, T., Vos, P. and Uknes, S. 1997. The Arabidopsis thaliana NIM1 protein shows homology to the mammalian transcription factor inhibitor I-kappa-B. Plant Cell 9:425-439.
Hunt, M.D., Delaney, T.P., Dietrich, R., Weymann, K., Dangl, J. and Ryals, J.A. (1997) Salicylate-independent lesion formation in Arabidopsis lsd mutants. Molec. Plant-Microbe Interact. 10: 531-16.
Lawton, K., Friedrich, L., Hunt, M., Weymann, K., Delaney, T.P., Kessmann, H., Staub, T. and Ryals, J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10: 71-82.
Delaney, T. P., Friedrich, L. and Ryals, J. 1995. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92: 6602-6606.
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E. and Ryals, J. 1994. A central role of salicylic acid in plant disease resistance. Science 266: 1247-1250.
Dietrich, R., Delaney, T., Uknes, S., Ward, E., Ryals, J. and Dangl, J. 1994. Arabidopsis mutants simulating disease response. Cell 77: 565-577.
Delaney, T. P., Friedrich, L., Kessmann, H., Uknes, S., Vernooij, B., Ward, E., Weymann, K. and Ryals, J. 1994. The molecular biology of systemic acquired resistance. In: Advances in Molecular Genetics of Plant-Microbe Interactions, Vol. 3, M. J. Daniels, J. A. Downie and Anne E. Osbourn, (eds.). Kluwer Academic Publishers, Dordrecht, Netherlands, pp. 339-347.
Uknes, S., Winter, A., Delaney, T., Vernooij, B., Morse, A., Friedrich, L., Nye, G., Potter, S., Ward, E. and Ryals, J. 1993. Biological induction of systemic acquired resistance in Arabidopsis, Molec. Plant Microbe Interact. 6: 692-698.
Updated November 2011
Questions or comments: terrence.delaney@uvm.edu
Home: http://www.uvm.edu/~tpdelane/lab/