Non-coding
DNA: non-repetitive "junk" DNA
| This
class of DNA is also known as selfish, ignorant,
parasitic and incidental DNA. It makes
up approximately 97% of the human genome. |
 |
"junk DNA".
A A general term that once encompassed all non-coding DNA when the
function was not understood. Dr. Susumu Ohno, writing in the Brookhaven
Symposium on Biology in 1972 in the article "Why So Much ‘Junk DNA" in
our Genome?" is credited with originating the term. But his paper was focused
"mainly on the fossilized genes, called pseudo genes, that are strewn like
tombstones throughout our DNA. As the term caught on in the 1980’s, its
meaning was extended to all non- coding sequences, the vast stretches of
DNA that are not genes and do not produce proteins" (about 95% of the genome).
In the last 2 decades however,
much has been learned and we know now that this "junk" contains many different
types of DNA sequences. "I don't think
people take the term very seriously anymore" says Eric Green [NHGRI] whose
group is mapping chromosome 7. [B. Kuska "Should
Scientists Scrap the Notion of Junk DNA?" JNCI 90(14): 1032-1033 July 15
1998]
Still, the term junk DNA is
frequently used incorrectly. Numerous articles in the medical
literature use junk and non- coding DNA interchangeably. [B. Kuska "Bring
in Da Noise, Bring in Da Junk" JNCI 90(15): 1125-1127 Aug. 5, 1998]
introns: everybody
knows what these are!!!
UTRs. The parts
of the messenger RNA sequence that do not code for product, i.e. the 5'
UNTRANSLATED REGIONS and 3' UNTRANSLATED REGIONS.
http://www.ebi.ac.uk/embl/Documentation/FT_definitions/feature_table.html

Non-coding
DNA: repetitive
sequences
| Non-coding
regions make up approximately 97% of the
human genome .......... Repetitive
Sequences make up at least 50% of the
genome. |
 |
Repetitive sequences
are thought to have no direct functions, but they shed light on chromosome
structure and dynamics. They hold important clues about evolutionary events,
help chart mutation rates, and by seeding DNA rearrangements, they can
modify genes and create new ones. They also serve as tools for genetic
studies.
The vast majority of repeated
sequences in the human genome are derived from transposable
elements - sequences like those that form
viral genomes - that propagate by inserting fresh copies of themselves
in random places in the genome. A full 45% of the human genome derives
from such transposable elements. A major surprise of this new global analysis
of the human genome is that many components in this diverse array of repeated
sequences, traditionally considered to be "junk," appear to have played
a beneficial role over the course of human evolution.
[NHGRI
"Summary of the Initial Sequencing and Analysis of the Human Genome" press
release, Feb. 11, 2001]
http://www.nhgri.nih.gov/NEWS/summary_of_sequence.html
Another important,
although much smaller, class of non-coding DNA are the tandem
repeats or satellite
sequences. These are associated with centromeres and telomeres, so they
may have a function, which is so far not understood.
1.
Transposable elements
-
Transposons
(move as DNA)
-
Retroposons
(Retrotransposons) (move as RNA intermediates)
-
Viral-like
Retroposons (terminal LTRs)
-
Nonviral-like
Retroposons (lack LTRs).
-
LINEs
(Long Interspersed Nuclear Elements)
-
SINEs
(Short Interspersed Nuclear Elements)
|
These are discrete
sequences in the genome that are mobile - they are able to transport themselves
to other locations within the genome.
-
The mark of a transposable element
is that it does not utilize an independent form of element
(such as phage or plasmid DNA). It can move itself alone and independently
from one site in the genome to another.
-
Unlike most other processes
involved in genome restructuring, transposition does not rely on any
relationship between the sequences at the donor and recipient sites
!!!!!
-
Transposable elements sometimes
incorporate additional sequences, and move them to new sites elsewhere
within the same genome. They are therefore an internal counterpart
to the vectors that can transport sequences from one genome to another.
They may provide the major source of mutations in the genome by:
-
insertion and interruption of
a functional gene
-
up-regulating or down-regulating
a gene as it is moved about the genome.
There are 2 types
of Transposable Elements:
-
Transposons
move directly as DNA. They do not go
through an RNA intermediate or reverse transcription. Instead the
enzyme transposase
is used to nick the host DNA and insert the transposon DNA into the cut
ends.
-
Retroposons
(retrotransposons) move through an
RNA intermediate
which then invades a DNA double helix at some point. The enzyme reverse
transcriptase then copies the retroposon into
DNA which is inserted into the DNA genome. There are 2
sub-types of Retroposons.
1.) viral-like
retroposons
which have LTRs (long terminal repeats).
2.) nonviral-like
retroposons which lack LTRs
|
Characteristics of Transposable
Elements
| Transposable Element |
Length |
Number/haploid genome* |
Fraction |
| Transposons |
2-3 Kb |
~ 300,000 |
3% |
| Viral-like Retroposons (with
LTRs) |
1-11 Kb |
~ 450,000 |
8% |
| LINEs (nonviral-like
retroposons; no LTRs) |
6-8 Kb |
~ 600,000 |
15-20% |
| SINEs (nonviral-like
retroposons; no LTRs) |
< 0.3 Kb |
~ 1 - 1,500,000 |
10-15% |
*
These estimates vary a great
deal depending on the reference!
|
| Transposons:
move directly as DNA without going through an RNA
intermediate or reverse transcription. The P
element of Drosophila, and the
Ac
element and
Ds
element in maize, are examples.
In replicative transposition,
the element is duplicated during the reaction. One copy remains at the
original site, while the other inserts at the new site. So transposition
is accompanied by an increase in the number of copies of the transposon.
In nonreplicative transposition,
the transposing element moves as a physical entity directly from one site
to another, and is conserved. This causes the element to be inserted at
the target site and lost from the donor site.
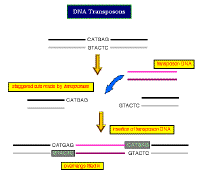 In
both processes, the critical enzyme is transposase
which is used to: In
both processes, the critical enzyme is transposase
which is used to:
-
nick the host DNA
-
insert the transposon DNA into
the cut ends.
Note:
this process does not depend upon complimentarity between the genomic and
host DNA!! It is therefore quite different from any mechanisms based
on recombination. |
Retroposons
(retrotransposons):
Retroposons
(retrotransposons) are related to retroviruses; their mobility
is based on the formation of an RNA intermediate which is then reverse
transcribed. The DNA copies then become integrated at new sites
in the genome. Transposition therefore involves an obligatory
intermediate of RNA. A diagnostic feature of retroposons
is the
generation of short direct repeats of target
DNA at the site of an insertion.
The difference between retroviruses and retroposons is
that the former have the ability to insert into a host genome and then
migrate
to other cells. The retroposon has only the ability to insert
into the host genome, although over the course of many generations, this
may occur many times.
Retroposons fall into two general classes:
1.) viral-like retroposons; 2.) nonviral-like retroposons.
Both types move through an RNA intermediate which then invades a DNA double
helix at some point. The enzyme reverse
transcriptase copies the retroposon into DNA
which is inserted into the DNA genome by integrase.
-
Members of the viral
superfamily. They
contain LTRs and code for reverse transcriptase and integrase
activities.
 They
are characterized by ~ 250- to 600 bp long terminal repeats (LTRs) flanking
the central protein-coding region as in retroviruses. Well known
examples are the Ty
elements
in yeast and copia
elements
in Drosophila. The mechanism
of transposition is similar to the mechanism of insertion for the retroviruses. They
are characterized by ~ 250- to 600 bp long terminal repeats (LTRs) flanking
the central protein-coding region as in retroviruses. Well known
examples are the Ty
elements
in yeast and copia
elements
in Drosophila. The mechanism
of transposition is similar to the mechanism of insertion for the retroviruses.
Note:
this process does not depend upon complimentarity between the genomic and
host DNA!! It is therefore quite different from any mechanisms based
on recombination.
|
-
Members of the nonviral
superfamily. These
elements lack LTRs. They may code for reverse transcriptase
and integrase activities.
-
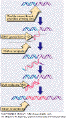 LINES
(Long
Interspersed Nuclear Elements) are long (average length = 6 500 bp),
moderately repetitive (about 10,000 copies). They encode a gene for the
enzyme transposase,
which is essential for their movement. LINEs are cDNA copies of functional
genes present in the same genome. LINES
(Long
Interspersed Nuclear Elements) are long (average length = 6 500 bp),
moderately repetitive (about 10,000 copies). They encode a gene for the
enzyme transposase,
which is essential for their movement. LINEs are cDNA copies of functional
genes present in the same genome.
-
Processed pseudo- genes are
a type of LINE which lacks a functional gene for transposase or any promotors.
They have the properties of an mRNA transcript (5' UTR; spliced
exons; short AT region derived from polyA tail). It is thought that
these pseudogenes originated by reverse transcription of a functional mRNA
which was randomly integrated into the germ cell of an ancient ancestor.
They are probably not pseudogenes which originated by duplication of whole
genes because they lack introns, have the remnants of a poly-a tail, and
do not have flanking sequences similar to those of the functional gene
copies. Because they are non-functional, they generally contain multiple
mutations, which are thought to have accumulated since their mRNAs were
integrated into chromosomal DNA.
-
SINES (Short
Interspersed Nuclear Elements). These are families of short
(150 to 300 bp), moderately repetitive elements of eukaryotes, occurring
about 1.000,000 - 1,500,000 times in a genome. Although they
possess external and internal features that suggest that they originated
in cellular transcripts (as opposed to retroviral sequences), they do
not
code
for proteins that have transposition functions. They appear to
be "dead" and incapable of transposition.
SINES appear to be DNA copies
of certain tRNA molecules, created presumably by the unintended action
of reverse transcriptase during retroviral infection.
-
Alu
elements, which occur on average every 6 Kb, are the most numerous SINES
in the human genome. Its short length and high degree of repetition
make it comparable to satellite DNA, except that the individual members
of the family are dispersed around the genome instead of being confined
to tandem clusters.
|
| There are few currently active
transposons in the human genome, but by contrast several active transposons
are known in the mouse genome. This explains the fact that spontaneous
mutations caused by LINES insertions occur at a rate of ~3% in mouse,
but only 0.1% in man. There appear
to be ~10-50 active LINES elements in the human genome. Neither
DNA transposons nor retroviral-like retroposons seem to have been active
in the human genome for 40-50 million years, but several examples of both
are found in the mouse. |

2.
Tandem Repeats:
These regions
consist of relatively short consensus sequences
which are repeated many times.
Rather than being interspersed throughout
the genome - like transposons,, the repeats occur one right after another
(in tandem).
There are 3 types of tandem
repeat elements - based roughly on size - but the terminology is not
precisely defined!
-
Satellite DNA is the highly
repetitive fraction of the genome consists of multiple
tandem copies of very short repeating units. The length of each
repeat unit is ~ 10 - 100 bp, but there may be 1,000 units or
more giving a total length of ~ 100 Kb. These often
have unusual properties:
-
They have an unusually high G-C
base content ratio, and may therefore be identified as a separate peak
from single copy DNA on a density gradient analysis. This gave rise
to the name satellite DNA.
-
They are often associated with
inert regions of the chromosomes, and in particular with centromeres.
-
In addition to the satellite sequences,
there are shorter stretches of DNA that show similar behavior, called minisatellites.
The length of minisatellite repeating units is ~10-100 bp. made
up of 20 - 50 units with a total length of 1-5 Kb.
-
Minisatellites are also
called VNTRs
(variable number tandem repeat) regions. Minisatellites undergo the same
sort of unequal crossing-over
between repeats They are useful in showing a high degree of divergence
between individual genomes that can be used for mapping purposes
or DNA fingerprints.
-
The name microsatellite
is usually used when the length of the repeating unit is <10 bp,
but many times they are dinucleotide repeats. Microsatellites
may undergo intrastrand mispairing, when slippage
of the polymerase occurs during replication
and this leads to expansion of the repeat.

|