|
|
Information is available
on:
Summary of Query Options
Browse lists
Summary display format
Selecting records and saving as text
LocusLink Reports
 Click
to see a sample LocusLink Report! Click
to see a sample LocusLink Report!

Summary of Query options
|
 |
The LocusLink query searches for any word (or word stem)
in a LocusLink report. Proximity searches, searches
on phrases, and synonyms are not supported ........ entering
serine
proteinase will retrieve records containing both serine and
proteinase,
but not necessarily serine proteinase.
| wild card |
* |
Field restriction
lower case, no space allowed between the term and the brackets;
the combination of wild card and field restriction supported only with
symbol[sym] |
[chr] chromosome |
| [loc] LocusID |
| [mim] MIM number |
| [sym] symbol |
| [pm] PubMed id |
| [ngi] nucleotide gi |
| [pgi] protein gi |
| booleans |
if not entered, and is implied with multiple words |
| no case sensitivity |
AND/and, OR/or, NOT/not are all supported |
| Note: Grouping by parentheses is not supported and organism-specific
restrictions are managed by the Organism: pull down menu. However,
the order in which query terms are added does make a difference. Query
terms are processed in order from left to right, and the next qery term
on the right is applied to the result of the previous operation. See also:
Constructing
URLs to LocusLink |
Examples of LocusLink Queries
| Query |
Result |
| A2M |
Browse list including A2M and IGHA2.
Click on the LocusID value (2, 3494 respectively) to display each LocusLink
Report. |
| A2* |
Browse list of loci with words beginning
with "A2" in any data field. |
| macroglobulin |
Browse list of loci with the word
"macroglobulin" in any data field. |
| 2[chr] |
Browse list of loci on chromosome
2. |
| protein kinase 3[chr] |
Browse list of loci with records
containing the word protein and the word kinase and a location on chromosome
3 |
| 12p1* |
Browse list of loci recorded as
having the upper range somewhere in 12p13, 12p12, or 12p11.
Note: This is a "string" search and not a range search.
Warning: Except for the field [sym], wild
cards cannot be used in combination with field-restriction (e.g. [mim],
[chr]) |
|
Browsing the Lists from a LocusLink Search
|
 |
The lists may be displayed in "Brief"
format..
| Data Element |
Definition 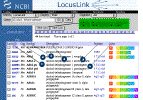 |
| LocusID (1) |
NCBI assigns a unique, stable LocusID
to each locus. Click on the number to retrieve the detailed LocusLink Report
page.
Note: As part of NCBI's Genome
Annotation Project, some LocusLink records may be generated which are
likely to be temporary and which therefore are not represented by a stable
ID. Instead, each is indicated by More, which is also to be clicked
to display the Report page. |
| Org (2) |
A two-letter abbreviation for the
organism in which this locus is described. The symbols (Hs for Homo
sapiens, Mm for Mus musculus, Rn for Rattus
norvegicus, Dr for Danio rerio, and Dm for Drosophila
melanogaster match the abbreviations used by UniGene. |
| Symbol (3) |
The official or alias symbol. Aliases
point to the official symbol. In some cases, a symbol may be an alias for
multiple loci. |
| Description (4) |
The gene name. |
| Position (5) |
The cytogenetic (human or rat) or
genetic (mouse) location. |
| Links (6) |
When links are available for PubMed,
OMIM, RefSeq, GenBank, HomoloGene, UniGene and variation data, they are
indicated as small color-coded blocks. Note: Neither PubMed nor GenBank
links are comprehensive. Use the Related Articles or nucleotide
neighbors functions to retrieve more records. |
Selecting records; Saving the record as text; Viewing the reports.
|
 |
If a query results in more than one record, you may select
to:
-
Save a subset of those records by checking the box to left of the LocusID
and then pressing Save Loci to provide a text display of the
reports. The text is formated as the LL_tmpl file available for
ftp.
See the
README
for a description of this format.
-
See a subset of those records by checking the box to left of the
LocusID and then pressing View Loci to generate LocusLink
Reports for those records.
LocusLink Reports 
|
 |
The query results are returned as a summary alphabetized
by Symbol (browse list). Any detailed LocusLink report
page is then accessed by clicking on the LocusID number at the left. Color-coded
icons support rapid jumps to related records in  ubMed, ubMed,  MIM, MIM, efSeq, efSeq,  GenBank
nucleotide, GenBank
nucleotide,  rotein, rotein,  omoloGene, omoloGene,  niGene,
and niGene,
and  ariation
data (dbSNP) ariation
data (dbSNP)
Seelecting the record to display.
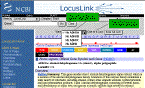 Although
multiple records can be selected for a report, only one record is displayed
in the browser at a time. The table of contents for the record is provided
in the blue column at the left, and the menu selection box named
view
allows you to switch displays.
The loci presented on the browse list are linked to a more detailed
report page. A report page may include the following information:
| Data Element |
Definition |
| Genomic
diagrams

|
The exon/intron organization of
the longest mature mRNA from genes and models aligned to genomic sequence
(NT_000000 accessions) is diagrammed (5'->3') at the top of each report.
Clicking on that display opens a new window showing alignments of non-model
RNAs to an NT_000000 accession. An option to see more details, via the
Evidence
Viewer, is provided via the option "Go to full display with alignments". |
| Button
Links |
Buttons are provided at the top
of each report page to indicate when information may be available from
a subset of other resources. (More links may be listed at the end of the
report page in the Additional
Links section.)
| Links to sites with more information |
 Cancer Gene Anatomy Project Tumor Suppressor and Oncogene Directory
Cancer Gene Anatomy Project Tumor Suppressor and Oncogene Directory
 Ensembl
Ensembl
 FlyBase
FlyBase
 The Genome Database
The Genome Database
 Human Gene Mutation Database
Human Gene Mutation Database
 HomoloGene
HomoloGene
 MapViewer
MapViewer
 Mouse Genome Database
Mouse Genome Database
 Online Mendelian Inheritance in Man
Online Mendelian Inheritance in Man
 Proteome, Inc.
Proteome, Inc.
 PubMed Publications
PubMed Publications
 RATMAP
RATMAP
 UCSC
Human Genome Project Working Draft UCSC
Human Genome Project Working Draft
 UniGene UniGene
 dbSNP Variation Database
dbSNP Variation Database
 Zebrafish Information Network
Zebrafish Information Network |
|
|
| Other links [more...] |
mv link
to MapView |
|
pm link to
PubMed for a specific concept |
|
sv link
to Sequence
Viewer |
 |
|
| Nomenclature [more...] |
Official Gene Symbol. The
official symbol and full name of a gene assigned by the genome-specific
nomenclature
committee. |
|
Interim Gene
Symbol. If the official symbol and name have NOT been
established, an interim symbol and name are provided. |
|
LocusID.
The unique NCBI LocusID assigned to each locus. LocusIDs are stable, genome-independent
identifiers. |
|
|
|
|
RefSeq.
Summary written by staff of the NCBI RefSeq group describing the function,
localization, and sequence properties of the gene and its products. |
|
Contributed Summary.
Summary supplied by non-RefSeq staff. |
|
Locus Type.
The category of the locus. Options include genes that encode proteins,
genes that encode untranslated RNA, mapped phenotypes, anonymous DNA segments,
models. |
|
Product.
The preferred gene product name. These names may be revised to reflect
current usage. |
|
Alternate Symbols These are
compiled from previous nomenclature, the published literature, or sequence
records. |
|
Alias. Alternative product
names . |
 |
|
|
|
This section describes the function
of the protein or RNA encoded by this locus. It may include Enzyme Commission
numbers, disease names established by OMIM, and/or terms from ontologies
such as Gene Ontology (GO).
When ontologies are displayed, the information table may contain:
-
the term
-
a coded representation of the evidence for this annotation
-
the source of the annotation
-
a link to PubMed
|
|
Whenever possible, the evidence codes are those
established by GO.
| Code |
Explanation |
| IMP |
inferred from mutant phenotype |
| IGI |
inferred from genetic interaction |
| IPI |
inferred from physical interaction |
| ISS |
inferred from sequence or structural similarity |
| IDA |
inferred from direct assay |
| IEP |
inferred from expression pattern |
| IEA |
inferred from electronic annotation |
| TAS |
traceable author statement |
| NAS |
non-traceable author statement |
| NR |
not recorded |
| E |
experimental evidence |
| P |
predicted/computed |
|
|
EC Number.
Enzyme commission number(s).. |
|
Phenotype.
The name of the disease that may result from variants at this locus; this
is linked to the OMIM record for the disease. |
|
|
|
|
This section is under development.
At present it may report human/mouse homologies and transcripts annotated
on the genome (models) that are related to known genes. |
|
Mouse/Human
Homology Map. links to NCBI's Human-Mouse
Homology Map which represents homologies that have been curated or
computed by sequence homology. The link on the symbol goes to the LocusLink
record of the homology, the Mm and
Hs
links go to the indicated map where gene order is based on mouse or human
coordinates, respectively. |
 |
|
|
|
Chromosome.
The chromosome(s) to which this locus has been mapped. |
|
Cytogenetic.
The cytogenetic position. |
|
Genetic.
The genetic position. |
|
Markers.
STS markers determined to be associated with this locus. These associations
are established either (1) computationally by ePCR with mRNAs associated
with this locus_id, (3)computationally from the NCBI's Genome Annotation
based on STS lying within the region defined by a LocusID, or (3) in a
published citation. If the STS is mapped, that location is also given.
The marker name links to dbSTS reports.
Markers mv:
link to STS on Map
Viewer
pm: link to
PubMed for paper providing details about the relationship between this
locus and this marker |
|
|
| NCBI Reference Sequences (RefSeq) |
|
All RefSeq records created for a
given locus are listed. Multiple records are distinguished by the brief
description of the transcript variant. [more...]
and [RefSeq
FAQ]
This section provides links to:
-
the RefSeq nucleotide record (genomic and mRNA accessions have 'NG_' and
'NM_' prefixes, respectively)
-
the RefSeq protein record (the 'NP_' prefix)
-
the CD browser report for each domain found in the RefSeq
-
the GenBank source sequence(s) used to construct the RefSeq record
-
BL provides a
quick link to the BLink
result for the protein.
In addition to providing links to the sequence and for contigs and models,
the
genome
annotation section also provides these links:
-
sv to the graphic
display mode of the NCBI sequence viewer
ev to the
graphic display (evidence viewer) of mRNAs aligning to the NT_ accesssion
in the region of the gene, with reports of mismatches, insertions, and
deletions relative to that genomic sequence for accessions not derived
from the NT_ accession (namely NM_ RefSeqs and GenBank accessions)
|
 |
|
|
|
A table of a subset of representative nucleotide and protein accessions
for the locus. These accessions are initially derived from a variety of
collaborations.
These data are included in our review processes and the accessions listed
for a given locus may change over time. EST accession numbers are provided
if no other sequence data are available to represent the locus. |
|
Type. Molecule type
for the nucleotide record.
-
m mRNA
-
g genomic DNA
-
e EST
u undetermined
|
|
Protein. accessions are linked
to Entrez sequence displays. BL
provides a quick link to the BLink
result for the protein. |
|
Strain. data, as annotated on the
GenBank record, is displayed when available. |
|
|
|
|
This section names and provides links to additional sites
that may contain information related to this locus.OMIM The MIM number
assigned to this gene product.UniGene The UniGene cluster that represents
this locus. |
|
OMIM. The MIM number assigned
to this gene product. |
|
UniGene. The UniGene cluster
that represents this locus. |
|