|
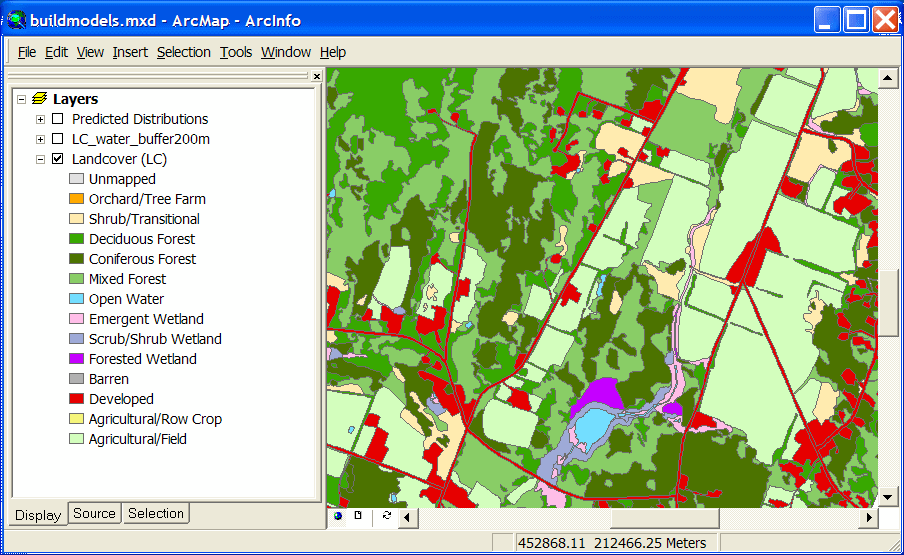
One common way to create predicted distribution maps for animals is to reclassify a land cover layer according to whether or not the species in question would be predicted to use each cover type. These simple species maps are often modified by using range data or such ancillary information as elevation or distance to water. The Spatial Analysis Lab has done several projects using this type of technique. Here is one example using a polygon- based land cover map developed for a single town (shown in the first graphic). |

A predicted distribution map for Eastern Kingbird (crosshatch), a species of open
fields and non-forested wetlands:
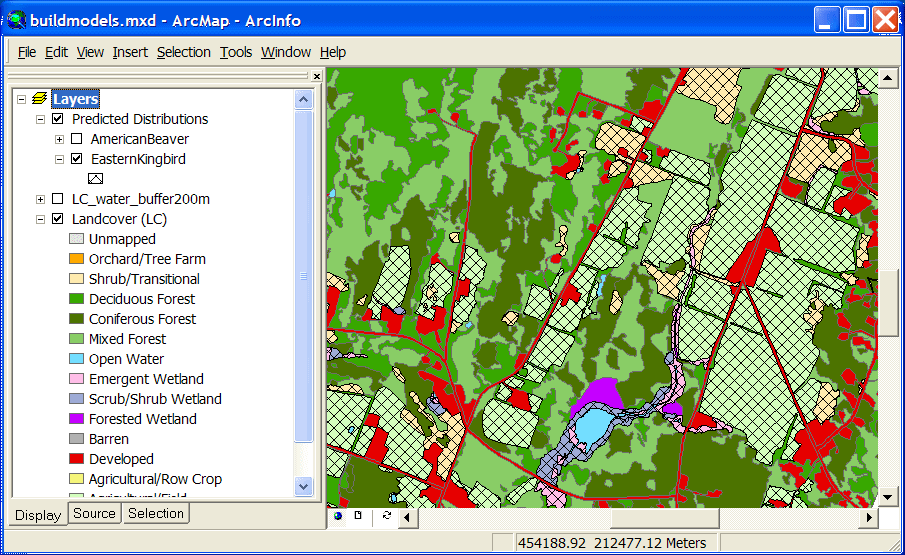
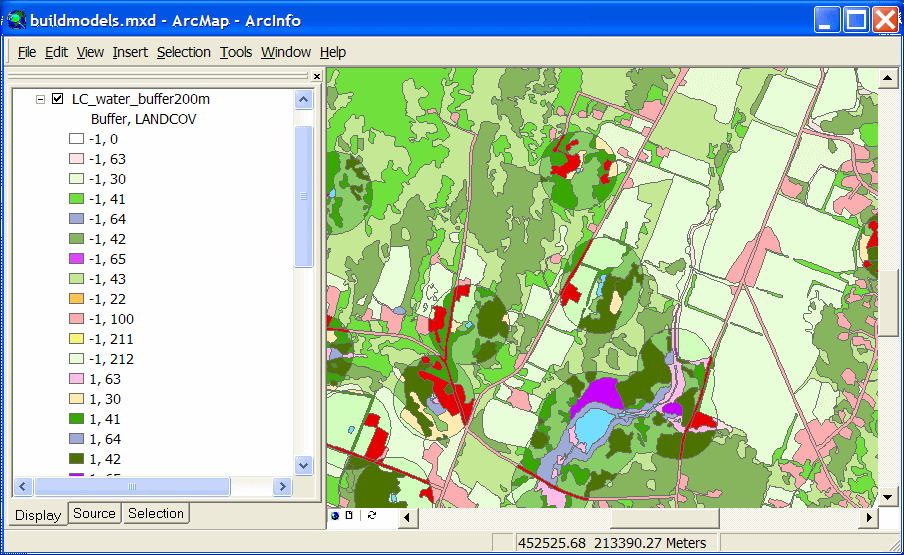
The land cover layer shown below has been modified by buffering the Open Water type and
overlaying it with the original land cover.
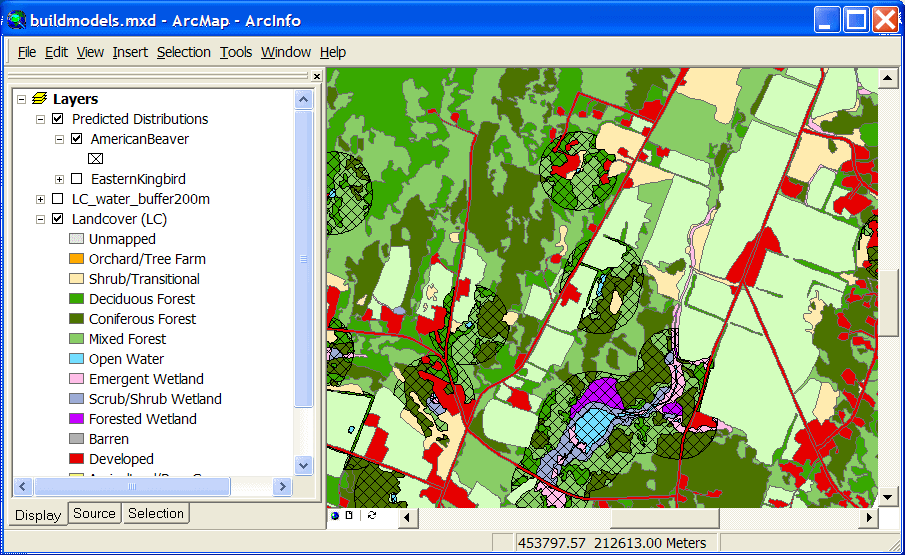
The modified land cover layer can then be used to create a predicted distribution map
for American Beaver (suitable cover types within 200 meters of water):
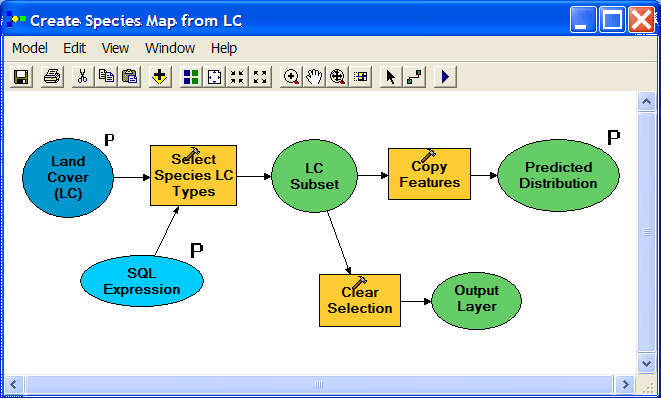
| A three-process model creates the simple distribution maps. Land cover types used by a species are selected using an SQL expression and are then copied to a new feature class. The selection is cleared in preparation for any subsequent work. Note that the input land cover layer, SQL expression, and desired output are all model parameters, which appear as choices for the user to specify when the model is run from Arc Toolbox. |
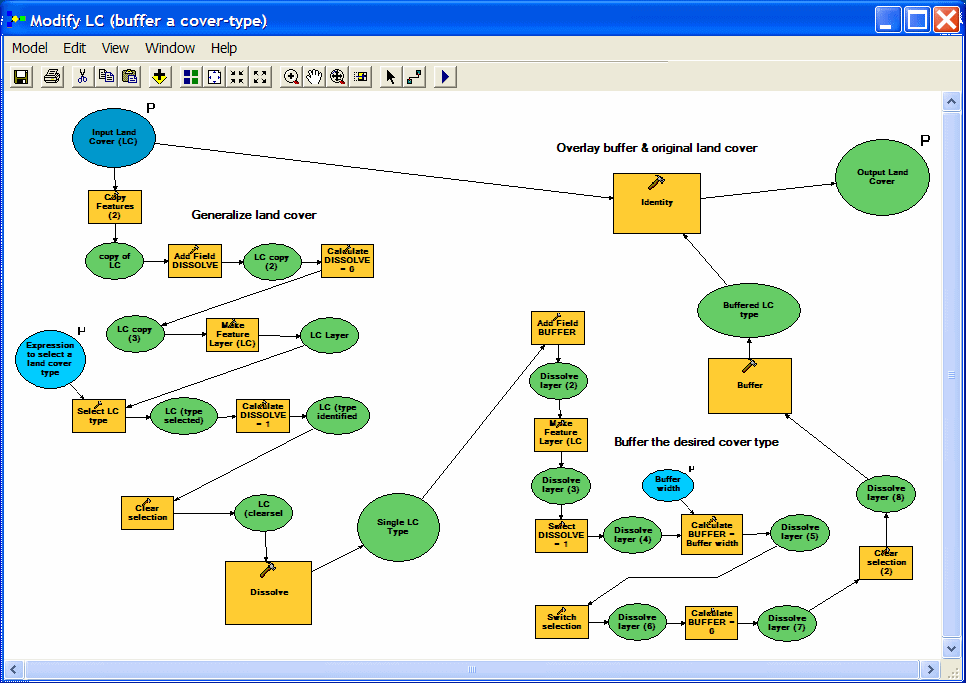
| A more complicated model was used to create a modified land cover layer (incorporating a buffer). The model is simpler than it looks: (1) input land cover is dissolved to isolate the class(es) that will be buffered, (2) the buffer operation is done, and (3) the buffer result is overlaid with the original land cover. Most of the extra steps are prep (adding items, calculating values....) for the dissolve and buffer processes. The buffer distance and SQL expression for selecting land cover type(s) tobe buffered are user inputs. |
A second example does similar modeling in a raster environment
and introduces use of Python scripting for automation.