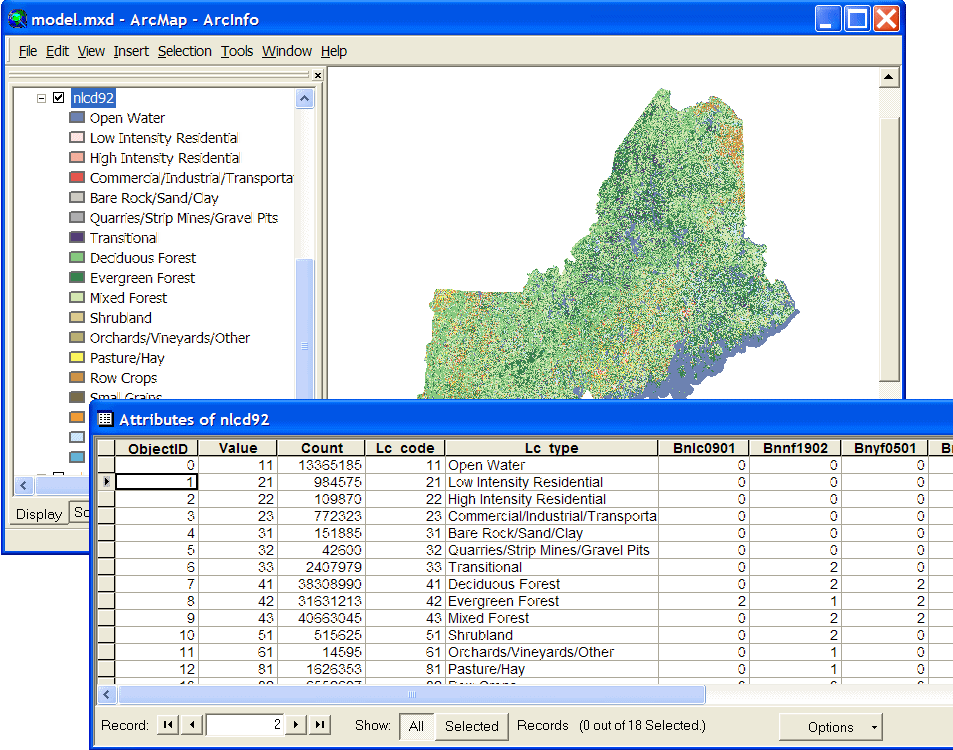
| This example originates with a project in the northern New England portion of Bird Conservation Region 14 (most of VT, NH, & ME), where we are using the 1992 National Land Cover Dataset (NLCD). We have added attributes to the NLCD Value Attribute Table to indicate which cover types are used by certain species. Field names beginning with "B" are species codes, and non-zero values indicate the cover type is used by the species. |

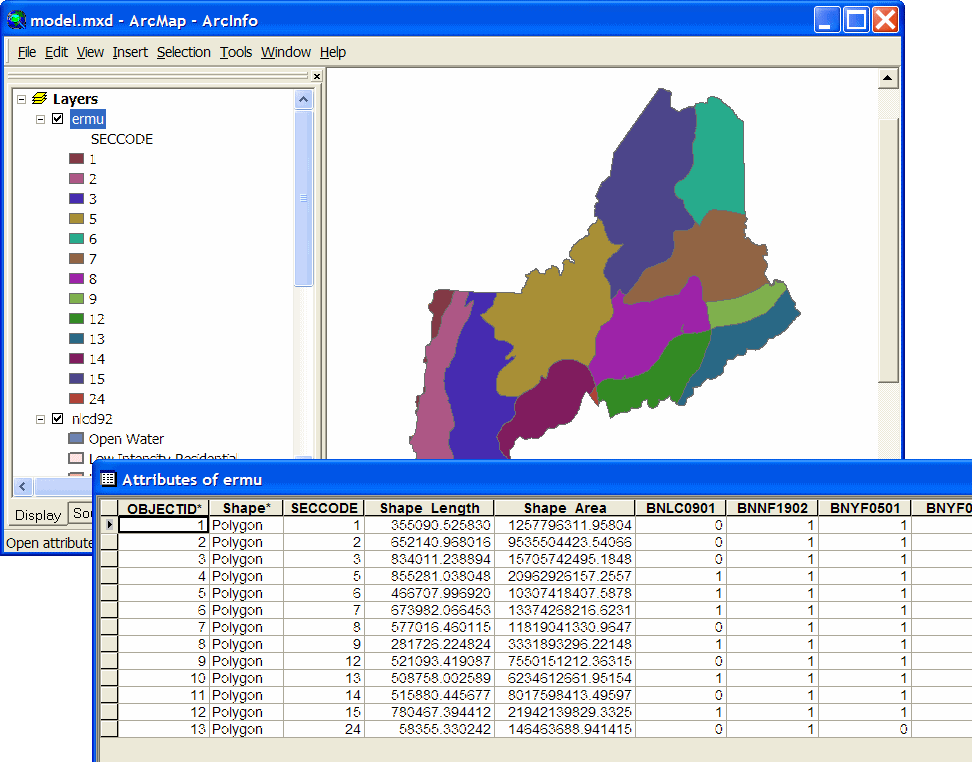
| As we are dealing with a larger area this time, we use species range data to refine the predicted distribution maps. In this case, ranges are represented by ecoregion (actually USFS "sub-section") polygons attributed with presence/absence data for each species. Here again, the extra attributes are species codes, and non-zero values indicate presence in the ecoregion. |
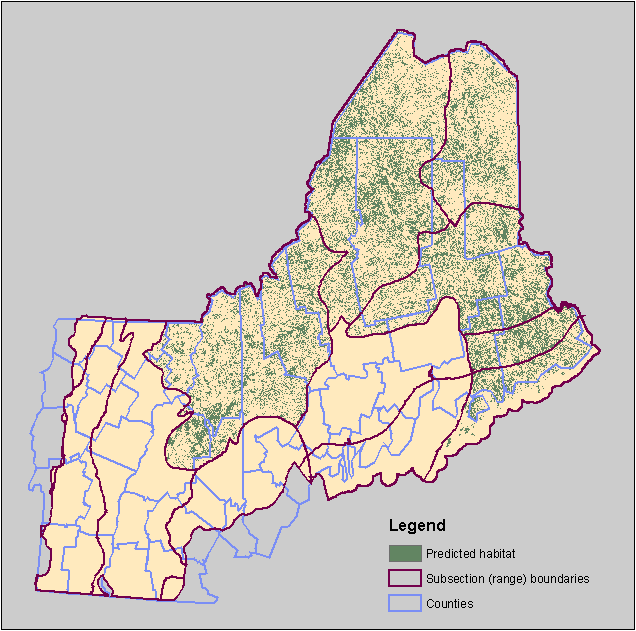
An end result looks like this predicted distribution map for Spruce Grouse:
| Three Spatial Analyst Tools go to work in this model. First, range polygons are converted to a binary raster layer (1 = species present; 0 = species absent). The second step reclassifies NLCD using a conditional statement to produce a second binary raster layer (1 = suitable cover; 0 = not suitable). Finally, the Map Algebra tool multiplies one binary raster layer by the other to produce a raster with value = 1 wherever both inputs are equal to 1 (appropriate cover type coinciding with species range). |
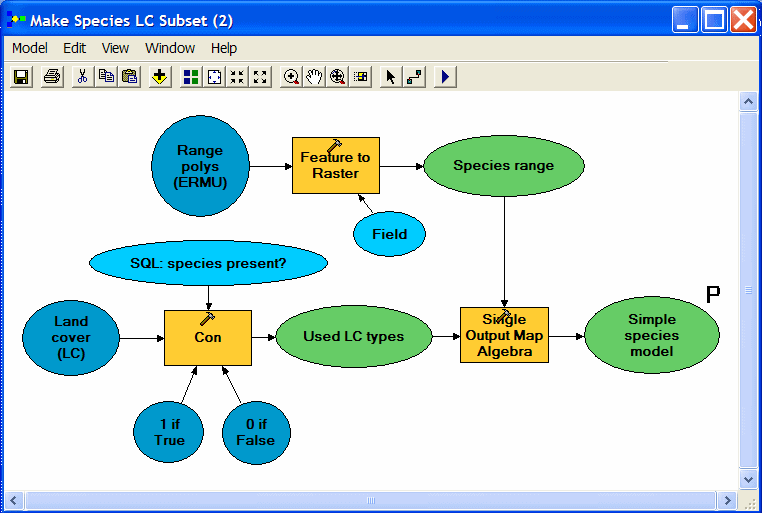
| It would be nice to run this model and have it produce maps for all our species by looping through each field (species code) in the data table. Although Model Builder is not yet up to this task, we can export to a Python script and add a few lines to make it loop. This export to script feature is also one way to begin to learn how Python scripting works. |
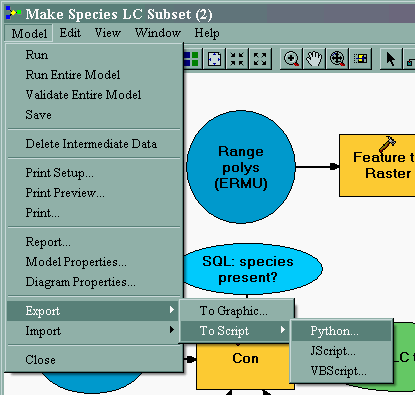
Here is the exported Python script just as ModelBuilder wrote it:
|
# --------------------------------------------------------------------------- # make_sp_map.py # Created on: Wed Jul 13 2005 11:05:25 AM # (generated by ArcGIS/ModelBuilder) # Usage: make_sp_map <Simple_species_model> # Description: # Creates a simple species model based on range data and land cover. Hard-coded # species ELCODE is used to rasterize ERMU to create a range map. Then land cover # (NLCD) is reclassed for the species and overlaid with the range raster. # --------------------------------------------------------------------------- # Import system modules import sys, string, os, win32com.client # Create the Geoprocessor object gp = win32com.client.Dispatch("esriGeoprocessing.GpDispatch.1") # Check out any necessary licenses gp.CheckOutExtension("spatial") # Load required toolboxes... gp.AddToolbox("C:/Program Files/ArcGIS/ArcToolbox/Toolboxes/Spatial Analyst Tools.tbx") gp.AddToolbox("C:/Program Files/ArcGIS/ArcToolbox/Toolboxes/Conversion Tools.tbx") |
|
# Script arguments... Simple_species_model = sys.argv[1] if Simple_species_model == '#': Simple_species_model = "D:\\species_maps\\bnlc0901simpl" # provide a default value if unspecified |
|
# Local variables... Species_range = "D:\\species_maps\\bnlc0901_lc" Used_LC_types = "D:\\species_maps\\con_nlcd921" Land_cover__LC_ = "nlcd92" v1_if_True = "1" v0_if_False = "0" SQL__species_present_ = "Bnlc0901 > 0" Range_polys__ERMU_ = "ermu" |
|
# Process: Feature to Raster... gp.FeatureToRaster_conversion(Range_polys__ERMU_, "BNLC0901", Species_range, "D:\\species_maps\\nlcd92") # Process: Con... gp.Con_sa(Land_cover__LC_, v1_if_True, Used_LC_types, v0_if_False, SQL__species_present_) # Process: Single Output Map Algebra... gp.SingleOutputMapAlgebra_sa("Sp_range * Used_LC_types", Simple_species_model, "D:\\species_maps\\bnlc0901_lc;D:\\species_maps\\con_nlcd921") |
|
The first (gray) section of code will be pretty much the same for any script exported
from ModelBuilder and can be ignored if you are already feeling code anxiety. The second
(red) code section sets a variable to the user-supplied argument (model parameter) for the
output dataset name. The green code section sets other local variables. These are simply
text strings in Python, and notice that their names match the names of the elements of the
model (see graphic above). The last (blue) code section is where the real work of running the 3 Spatial Analyst processes is done. Note that each process (tool) takes several
arguments, which are all text strings or string variables.
The script (and the model) create a map for a single species with code "BNLC0901". To make the script loop, we simply need to replace the species code argument with a variable and add some code to cycle this variable through the list of species codes. We have the necessary species codes as field names in one of our attribute tables (see above) and also in a different table (NLCD92.DAT) that we will use here. Shown below is the same script after modifications to make it loop. The code necessary for looping is highlighted in yellow. Note that some of the local variables (green section) have moved inside the while loop because they change with each new species code. |
|
# --------------------------------------------------------------------------- # model_loop.py # Created on: Fri Mar 11 2005 06:51:06 PM # (generated by ArcGIS/ModelBuilder & modified to allow looping) # Usage: model_loop # Description: # Creates a simple species model based on range data and land cover. # Species ELCODE (from field names in LC model table) is used to rasterize # ecoregions (ERMU) to create a range map for each species/loop, then land cover # (NLCD) is reclassed according to records for the species in the LC matrix # and overlaid with the range raster. # --------------------------------------------------------------------------- # Import system modules import sys, string, os, win32com.client # Create the Geoprocessor object gp = win32com.client.Dispatch("esriGeoprocessing.GpDispatch.1") # Check out any necessary licenses gp.CheckOutExtension("spatial") # Load required toolboxes... gp.AddToolbox("C:/Program Files/ArcGIS/ArcToolbox/Toolboxes/Spatial Analyst Tools.tbx") gp.AddToolbox("C:/Program Files/ArcGIS/ArcToolbox/Toolboxes/Conversion Tools.tbx") gp.AddToolbox("C:/Program Files/ArcGIS/ArcToolbox/Toolboxes/Data Management Tools.tbx") |
|
# Local variables... Land_cover = "D:/species_maps/nlcd92" v1_if_True = "1" v0_if_False = "0" Range_polys = "D:/species_maps/ranges.mdb/ermu" modelPath = "D:/species_maps/" # Set the workspace gp.Workspace = modelPath try: |
|
# Get list of fields (species codes) for looping fields = gp.ListFields("D:/species_maps/nlcd92.dat", "B*") field = fields.Next() while field: species = field.Name |
|
# Build output data layer names from species code: Sp_range = modelPath + species + "_rng" Used_LC_types = modelPath + species + "_lc" Simple_species_model = modelPath + species + "simpl" # SQL and map algebra expressions for later process steps: SQL__species_present_ = species + " > 0" InExpression = Simple_species_model + " = (" + Sp_range + " * " + Used_LC_types + ")" |
|
gp.AddMessage("Working on " + species; converting range polys to raster...." # PROCESS: Feature to Raster... gp.FeatureToRaster_conversion(Range_polys, species, Sp_range, "30") gp.AddMessage(" Reclassifying land cover....") # PROCESS: Con... gp.Con_sa(Land_cover, v1_if_True, Used_LC_types, v0_if_False, SQL__species_present_) gp.AddMessage(" Overlaying reclassified land cover with range....") # PROCESS: Multiple Output Map Algebra... gp.MultiOutputMapAlgebra_sa(InExpression) # Delete intermediate data layers: gp.AddMessage(" Cleaning up....") gp.Delete(Sp_range) gp.Delete(Used_LC_types) |
|
field = fields.Next() |
|
except: gp.AddMessage(gp.GetMessages(2)) print gp.GetMessages(2) |
|
Note also that we no longer have an input argument. Other changes to the script include
the addition of messages that will be displayed as the script is running and a try/except
block meant to help catch errors. (This is a very minimal use of Python's error handling
capabilities.) I've also written the paths in the simpler, UNIX-like format using a forward
slash (/) rather than the more cumbersome double backslash (\\) produced by ModelBuilder.
It's all the same to Python, so why waste characters?
For more information about geoprocessing scripting in ArcGIS, review the ArcGIS help installed with your software, visit the ESRI web site, or check out some of the digital books that came with ArcGIS. The while loop and other useful looping structures are explained in the ESRI document Writing Geoprocessing Scripts in ArcGIS, which should be installed in the "Documentation" folder of your ArcGIS installation folder. For all things Python, visit python.org . |
July 2005 . . . . questions/comments to: Ernest Buford