| |
Entrez Help Document |
| PubMed | Entrez | BLAST | OMIM | Books | Taxonomy | Structure |
Last modified: February 2006.
Nucleotide - Protein - Genome - Structure - PopSet
| Table of Contents |
| Introduction |
Entrez integrates the scientific literature, DNA and protein sequence databases, 3D protein structure and protein domain data, population study datasets, expression data, assemblies of complete genomes, and taxonomic information into a tightly interlinked system. Help using the literature component of Entrez, known as PubMed, is also available. Go to PubMed Help.
This help document is organized as follows:
Introduction - describes the Entrez cross-database search page, its databases, and features.
Searching - introduces and demonstrates basic search techniques.
Refining Your Search - demonstrates advanced search techniques using Limits, Preview/Index, and History and includes help with Writing Advanced Search Statements.
Displaying and Saving Results - explains the various display formats, how to save results, and how to link to related information in other databases.
LinkOut - introduces this newest Entrez feature and explains how to use it.
Entrez, the Life Sciences Search Engine
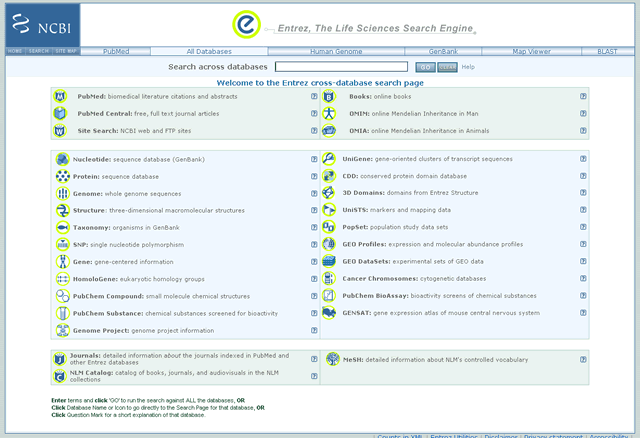
The Entrez page is home to the Entrez Global Query database search (the Entrez cross-database search page). It is available at: http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi. The entire group of individual Entrez databases is organized on this page with literature databases at the top including PubMed, PubMed Central, Journals, Books, and OMIM. The NCBI Site Search is also listed. The sequence databases include Nucleotide, Protein, Genome, Structure, and SNPs. The remaining databases are Taxonomy, Gene, UniGene, Conserved Domains, 3D Domains, UniSTS, PopSet, GEO Profiles, GEO Datasets, and Cancer Chromosomes. Links to popular NCBI Web pages, such as PubMed, Human Genome, Map Viewer, and BLAST, are on the toolbar. There is also a link to the "GenBank" database, which leads to the Nucleotide database.
By using the Entrez Global query, a search across all Entrez databases is performed by entering a simple search term or phrase in the "Search across databases" query box. Select the Go button to execute the search, or press the Enter button on your keyboard. The CLEAR button erases search terms in the query box; use it to begin a new search. The results found in each database are displayed on the Global Query page. Click on the result number or its adjacent database name to get to the specific results. See the link to the Global Query Help document, which is to the right of the CLEAR button.
| The Databases |
As an example of the change in the format of results returned from an Entrez Nucleotide search, consider a simple search using a query of "mouse[organism]". The result are shown with links to the split of data in the GenBank EST division, the GenBank GSS division, and the CoreNucleotide subset. Users can select results from the specific nucleotide dataset of interest by clicking on the appropriate results link.
The new component databases are included within the Entrez linking scheme and Links within and between databases can be selected as usual from the various datasets, Popular search strategies such as the Limits, Preview/Index, History, and MyNCBI can be used within each individual database.
Specialized search fields are available for the each new Nucleotide component database and can be seen in the respective indexed search fields in the "Add Term(s) to Query or View Index:" section of the Preview/Index tab for each. New fields available for the EST component database include EST ID, EST Name, and Library. Within the GSS component database, new fields include GSS ID, GSS Name, Library Class, and Library Name. CoreNucleotide contains the same 23 search fields as the traditional Nucleotide database.
All previous Entrez functionality remains, such as Clipboard where searches can be be saved temporarily and MyNCBI where searches can be indefinitely saved to run at user-selected intervals. Details and History pages are available for the individual search sets. To see search Details, the database of interest must be selected when performing a search from the main Nucleotide page.
Patent sequences are incorporated through arrangements with the U.S. Patent and Trademark Office (USPTO) and via the collaborating international databases from other international patent offices.
The Umbrella Nucleotide database also includes the Reference Sequence (RefSeq) records. RefSeqs are an NCBI-curated non-redundant set of sequences. Click on this link for more information about the RefSeq project.
Use Cn3D, the NCBI 3D structure viewer, for easy interactive visualization of molecular structures from Entrez.
Links between databases are also possible. Protein and Nucleotide neighbors are determined by performing similarity searches using the BLAST algorithm to compare the entry amino acid or DNA sequence to all other amino acid or DNA sequences in the database. Nucleotide sequence records in the Nucleotide database are linked to the PubMed citation of the article in which the sequences were published. Protein sequence records are linked to the nucleotide sequence from which the protein was translated.
See Displaying and Saving Results for more information on links within and between databases.
| Limits |
Limits allow restriction of a search to a defined subset of the database. Limits can be set to restrict a search to a particular database field (e.g., the Author field). Limits can be set to search everything but a particular type of data (e.g., “exclude patent records”). Alternatively, limits can be set to search only a particular type of data (e.g., Genomic RNA/DNA) or to search only data from a particular source database (e.g., EMBL). Date limits and sequence length limits are also possible.
The contents of each Entrez database differ, and therefore the Limits available for each database differ. See the “Limits Available by Database Summary” in the Summary Matrices section of this introduction. See also the Using Limits section of this document for help in using limits in your search.
Limits available for the Component Nucleotide and CoreNucleotide database
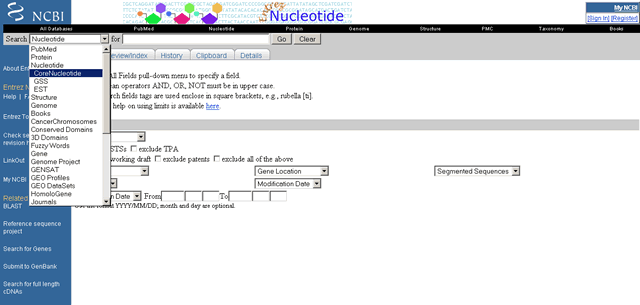
| Preview/Index |
Indexes are alphabetical lists of terms from searchable database fields. When indexes are displayed, they provide a way to browse the terms by which records and/or data are described. Entrez not only lets you browse indexes, you can also select terms to search directly from them.
As with limits, the indexes available for a particular database are dependent on the searchable fields of that database. See the “Indexes Available by Database” in the Summary Matrices section of this introduction.
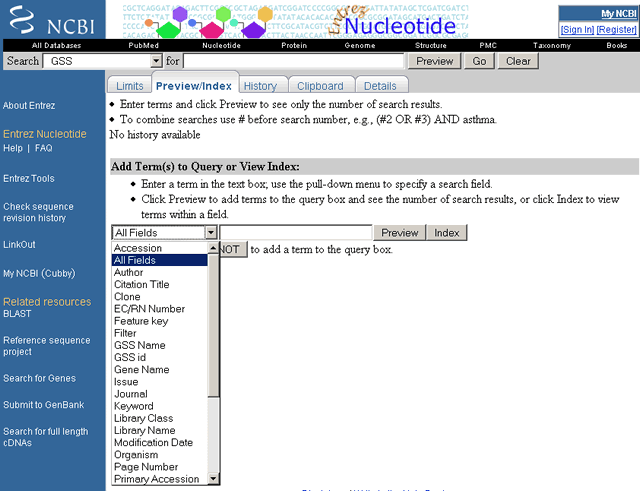
The view below displays the entries listed alphabetically under "bacter" in the Organism index of the Nucleotide database. Specific indexes are selected from the "Add Term(s) to Query or View Index" pull-down menu. Search by typing search terms in the query box and select the Index button. Browse the terms by selecting the Up and Down buttons to scroll. See the Using the Indexes section of this document for help in using indexes in your search.
Component Nucleotide Databases All Fields Index
Available indexes for the Nucleotide database are shown here in the Nucleotide "Add Terms to Query or View Index" pull-down menu
Available indexes for the Nucleotide database are shown below.
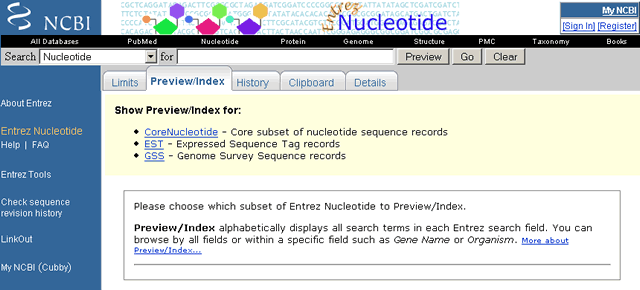
The Umbrella Nucleotide "Add Term(s) to Query or View Index" pull-down menu is shown
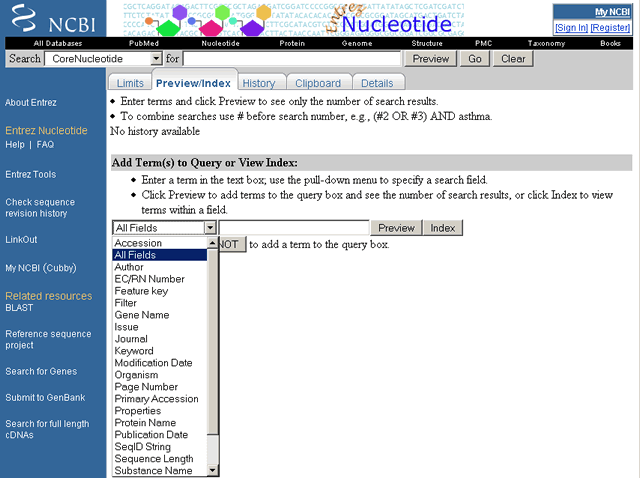
The Core Nucleotide "Add Term(s) to Query or View Index" pull-down menu is shown
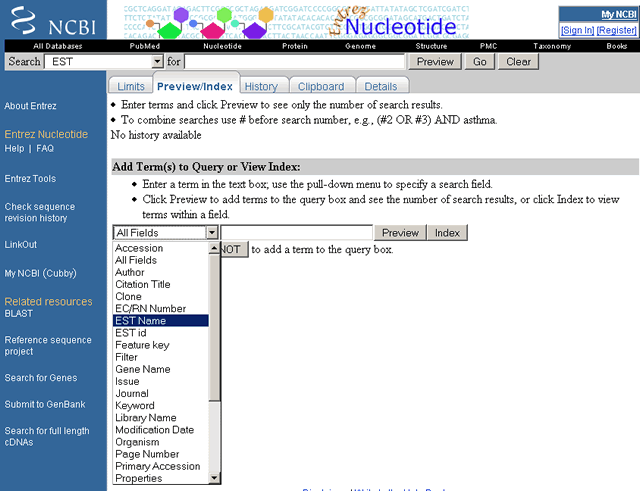
The EST Nucleotide "Add Term(s) to Query or View Index" pull-down menu is shown
The GSS Nucleotide "Add Term(s) to Query or View Index" pull-down menu is shown
| History |
Using the Preview option of Preview/Index allows a searcher to display the last three results for consecutive searches. A searcher can view the effect of each successive limit added to the search strategy. See the explanation of History in the next section for an option to see all search history for individual Entrez databases.
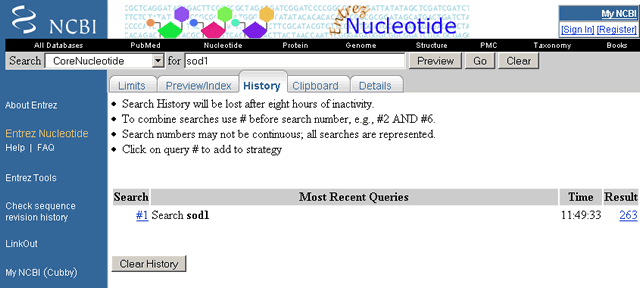
History provides a record of the searches performed during a search session. Histories are database specific. Each time search terms are typed into the query box and the search is executed, the search terms, the time the search was executed, and the search results are numbered consecutively and saved automatically in the History for that database. The History can be recalled at any time during a search session, but histories are lost after eight hours of inactivity. Use Histories to review, revise, or combine the results of earlier searches. See the Using Your History section of this document for help in using your search history.
Sample History page for CoreNucleotide
| Clipboard |
The Clipboard is a temporary place to save search results. Each Entrez database has its own. Search results are not saved automatically. Each database clipboard is limited to 500 items, and items saved to the clipboard are lost after 8 hours of inactivity. Items can be displayed and saved from the Clipboard. See the Details, Send To Clipboard, and Save section of this document for help in adding records to and using records on your clipboard.
| Searching |
Enter one or more search terms (e.g., 16S RNA) in the query box to search all databases or select one database, such as 'Nucleotide', and enter the query on the Home Page for that database.
Entrez Cross Database Search Engine
Sample Entrez Global Query Page
| Subject Searching |
Subject terms are automatically combined (ANDs). In the example above, the search query - 16S RNA - retrieves all records with the terms 16S AND RNA. See Boolean Operators for more information on combining terms with Boolean Operators.
| Phrase Searching |
To force Entrez to search for a phrase, enter double quotes (" ") around the phrase. For example, "16S RNA" retrieves fewer documents when compared with the subject search 16S AND RNA, which retrieves many more.
Using quotes forces Entrez to check a phrase list, against which the search terms are matched. It is not true adjacency searching. If the search phrase is not in the phrase list, Entrez returns an informational message "See Details.No items found." and once a nucleotide component database is selected, the message " Quoted phrase not found. See Details.No items found. .
Although phrase searching is useful, it should be used with caution because enclosing search terms in quotes restricts the documents retrieved to only those documents with exact matches to the text string within the quotes. Also, note that not all phrases will be indexed in Entrez nucleotide or protein indexes. Therefore, you may also wish to re-run the search without enclosing the terms in quotation marks. Carefully check the results to see if your phrase was found.In the "16S RNA" example, documents with the the phrase '16S RNA' are retrieved, but documents with the term 16S RNA gene are not retrieved. You can see the Pubmed help document for information on how PubMed handles quoted searches: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&db=books&doptcmdl=GenBookHL&term=quotes+AND+helppubmed%5Bbook%5D+AND+404082%5Buid%5D&rid=helppubmed.section.pubmedhelp.Searching_PubMed#pubmedhelp.Searching_for_a_phra
| Searching for Authors |
Enter author names in the format: last name plus initials (e.g., johnson d). Do not use punctuation. This format instructs Entrez to search only the Author field. Entrez automatically truncates on the author's name to account for various initials and designations, such as Jr. or 2nd. If only a last name is used in the query box (e.g., johnson), Entrez will search All Fields for that term.
| Searching for Unique Identifiers |
Unique identifiers can be accession numbers, which apply to a complete sequence record, or sequence identification numbers, which apply to the individual sequences within a record.
The format of accession numbers varies, depending upon the source database. (As noted above in The Databases section, each data domain in Entrez contains records from a number of different sources.) Some examples of typical accession number formats are below. The Sample GenBank Record contains additional detail about accession numbers.| Type of Record | Sample Accession Format |
| GenBank/EMBL/DDBJ Nucleotide Sequence Records |
One letter followed by five digits, e.g.: Two letters followed by six digits, e.g.: |
|
GenPept Sequence Records (which contain the amino acid translations from GenBank/EMBL/DDBJ records that have a coding region feature annotated on them) |
Three letters and five digits, e.g.: |
| Protein Sequence Records from SWISS-PROT and PIR |
All are six characters: e.g.: |
| Protein Sequence Records from PRF |
A series of digits (often six or seven) followed by a letter, e.g.: |
| RefSeq Nucleotide Sequence Records |
Two letters, an underscore bar, and six digits, e.g.: mRNA records (NM_*): genomic DNA contigs (NT_*): |
| RefSeq Protein Sequence Records |
Two letters (NP), an underscore bar, and six digits, e.g.: |
| RefSeq Model (predicted) Sequence Records from the Human Genome annotation process |
Two letters (XM, XP, or XR), an underscore bar, and six digits, e.g.: |
| Protein Structure Records |
PDB accessions generally contain one digit followed by three letters, e.g.: MMDB ID numbers generally contain four digits, e.g.: The record for the Tumor Suppressor P53 Complexed With DNA can be retrieved by either number above. |
| Searching by Molecular Weight |
002002 [Molecular Weight]or a range of weights:
002002:002009 [Molecular Weight]or either expression can be combined with other Entrez search terms, for example, to limit by organism:
002002:002009 [Molecular Weight] AND human [Organism]Note that molecular weight must be entered as a fixed six-digit field, filled with leading zeros (not letter O). The square brackets can contain the full spelling of the search field, as in the examples above, or the abbreviation [MOLWT] in upper- or lowercase. Note also that where cleavage products are annotated with features, the molecular weight of each cleavage product is calculated, not the molecular weight of the whole protein. Thus, you may retrieve a large protein when querying with a small molecular weight; be sure to check the feature table of the protein record to see if it has cleavage products. How the Molecular Weight is calculated:
| Range Searching |
Note: It is not possible to search for a range of sequence identification numbers (known as GI numbers and Version) numbers.Range of sequence lengths:
Note: Molecular weights must be expressed in six digits, filled with leading zeros (not letter O). Additional information about Searching by Molecular Weight is included above.Also, a range search can be combined with other Entrez search terms, for example, to limit by organism:
| Truncating |
Truncating search terms is a convenient way to find all the records that contain terms that begin with a given text string. Place an asterisk (*) at the end of a search term to find all records with a term that begins with that text string. For example, the truncated search term "immunoglob*" will retrieve all records in the database that contain the word immunoglobulin, immunoglobulins, immunoglobin, and immunoglobins.
Entrez searches the first 150 variations of a truncated term. If a truncated term produces more than 150 variations, which is possible with terms like "bact*," Entrez gives the following warning:
"Wildcard search for 'bact*' used only the first 150 variations. Lengthen the root word to search for all endings."
Phrases that include a space in the word after the asterisk will NOT be retrieved. For example, if you search "chromo*," the documents retrieved will contain terms like chromobacterium but not chromo helicase.
Left-handed truncation is not possible (e.g., "*bacterium").
| Combining Sets |
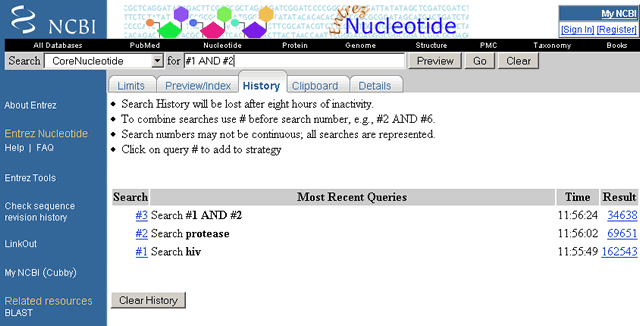
Use your search History to combine documents retrieved with different search terms at different times during your search session. For example, search the Nucleotide database for HIV. This search retrieves over 100,000 documents. Now search the Nucleotide database for protease. This search retrieves over 50,000 documents. Now click on the History for the Nucleotide database.
The results for the HIV and protease search terms are saved as Search Sets #1 and #2, respectively. In the query box, type #1 AND #2 and select Go. This search combines the documents in Search Set #1 (HIV) with the documents in Search Set #2 (protease) and retrieves only those documents that are in both sets.
Click on History again and note Search Set #3 (#1 AND #2).
Remember, this History is for the Nucleotide database only, and it will be lost after 8 hours of inactivity. See Boolean Operators and Using Your History for more information and examples.
Sample Search History in CoreNucleotide to Combine Search Terms
| Refining Your Search |
Sometimes it is necessary to refine your search statement by using the Limit, Preview/Index, and History options of a given Entrez database. The key to using these options, especially the Limit and Preview/Index options, is to better understand the search fields and Boolean Operators of the Entrez databases.
| Boolean Operators |
Boolean Operators used in Entrez are:
AND: To AND two search terms together instructs Entrez to find all documents that contain BOTH terms.
OR: To OR two search terms together instructs Entrez to find all documents that contain EITHER term.
NOT: To NOT two search terms together instructs Entrez to find all documents that contain search term 1 BUT NOT search term 2.
The Entrez search rules and syntax for using Boolean operators are:
1. Boolean operators AND, OR, NOT must be entered in UPPERCASE (e.g., promoters OR response elements).
2. Entrez processes all Boolean operators in a left-to-right sequence. The order in which Entrez processes a search statement can be changed by enclosing individual concepts in parentheses. The terms inside the parentheses are processed first as a unit and then incorporated into the overall strategy. For example, the search statement: g1p3 AND (response element OR promoter) is processed by Entrez by ORing the terms response element OR promoter first and then ANDing the resulting set of documents with g1p3.
3. Click on the Details button to see how Entrez translated and executed your search strategy.
4. See Writing Advanced Search Statements for more information on using Boolean Operators and Entrez Search Field Qualifiers.
| Using Limits |
Limits are used to refine search results to retrieve only the most relevant documents. In other words limits remove unneeded or unwanted documents. This section provides examples for using limits to:
Example: You are only interested in nucleotide sequences from the mouse:
1. Starting at the NCBI Home Page www.ncbi.nlm.nih.gov, select the Nucleotide database from the black menu bar or the Search pull-down menu.
2. Select Limits.
3. In the "Limited To:" section, select Organism from the Search Field pull-down menu.
4. Type "mouse" without quotes in the query box and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Field: Organism). You may also link to the results in CoreNucleotide, EST, or GSS.
Example: You are only interested in protein sequences that are less than 50 amino acids in length:
1. Select the Protein database from the black menu bar or the Search pull-down menu.
2. Select Limits.
3. In the "Limited To:" section, select Sequence Length from the Search Field pull-down menu.
4. Type "0:50" without quotes in the query box and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Field: Sequence Length).
Example: You are interested in mitochondrial carriers, but you do not want the EST sequences:
1. Select the CoreNucleotide database from the Search pull-down menu.
2. Type "mitochondrial carrier" without quotes in the query box.
3. In the "Limited To:" section, check the box next to "exclude ESTs" and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Limits: exclude ESTs).
In the Nucleotide database, you can exclude EST, STS, GSS, working drafts, and/or Patent sequences. In the Protein database, you can exclude Patent sequences.
Example: You are only interested in Cryptosporidium ribosomal RNA sequences:
1. Select the Nucleotide database from the black menu bar or the Search pull-down menu.
2. Type "cryptosporidium" without quotes in the query box.
3. Select Limits.
4. In the "Limited To:" section, select the "Molecule" pull-down menu and choose rRNA and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Limits: rRNA). You may also link to the results in CoreNucleotide, EST, or GSS.
Example: You are interested in the genes in the chloroplast of flowering plants:
1. Select the Nucleotide database from the black menu bar or the Search pull-down menu.
2. Type "flowering plants" without quotes in the query box.
3. Select Limits.
4. In the "Limited To:" section, select the "Gene Location" pull-down menu and choose Chloroplast and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Limits: Chloroplast). You may also link to a majority of the records in CoreNucleotide.
Example: You are interested in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. You know that there are several segmented sets of sequences associated with the CFTR gene. But you are only interested in displaying the master record of any segmented sets associated with the CFTR gene:
1. Select the Nucleotide database from the black menu bar or the Search pull-down menu.
2. Type "cftr" without quotes in the query box.
3. Select Limits.
4. In the "Limited To:" section, select the "Segmented Sequences" pull- down menu and choose "Show only master of set" and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Limits: Show only master of set).
Please note that this option does not allow you to limit the documents retrieved to only those containing segmented sequences. It simply allows you to control how segmented sets of sequences are displayed.
Example: You are interested only in cysteine phosphatase protein sequences submitted directly to PIR:
1. Select the Protein database from the black menu bar or the Search pull-down menu.
2. Type "cysteine phosphatase" without quotes in the query box.
3. Select Limits.
4. In the "Limited To:" section, select the "Only from" pull-down menu and choose PIR and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Limits: PIR).
1. Select the Nucleotide database from the black menu bar or the Search pull-down menu.
2. Type "pigs" without quotes in the query box.
3. Select Limits.
4. In the "Limited To:" section, select Organism from the Search Field pull-down menu.
5. In the "Limited To:" section, select the "Modification Date" pull-down menu, choose "30 Days", and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Field: Organism, Limits: 30 Days).
Example: You want to retrieve all mouse or human protein sequences added to the database (or updated) during 1997:
1. Select the Protein database from the black menu bar or the Search pull-down menu.
2. Select Limits.
3. Type "mouse OR human" without quotes in the query box.
4. Select Limits.
5. In the "Limited To:" section, select Organism from the Search Field pull-down menu.
6. In the "Limited To:" section, select the "Modification Date" pull-down menu, and choose Modification Date (as opposed to Publication Date). In the date boxes, type the dates in the format YYYY/MM/DD. You can tab from box to box in the date fields. The From date is 1997/01/01, and the To date is 1997/12/31. Select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Field: Organism, Limits: Modification Date, from 1997/01/01 to 1997/12/31).
Example: You are interested in the protein translations of human GenBank nucleotide sequences added to the protein database (or updated) in the last 30 days. You do not want patent records:
1. Select the Protein database from the black menu bar or the Search pull-down menu.
2. Select Limits.
3. Type "human" without quotes in the query box.
4. Select Limits.
5. In the "Limited To:" section, select Organism from the Search Field pull-down menu.
6. On the same screen, select the "exclude patents" check box, select GenBank from the "Only from" pull-down menu, and finally select "30 Days" from the Modification Date pull-down menu and select Go.
On the results screen, note that the check box next to Limits is checked, indicating that Limits are selected and active. Beneath the check box, the selected and active limits are highlighted in yellow (i.e., Field: Organism, Limits: Exclude patents, 30 Days, GenBank).
| Using the Indexes |
Indexes are used to browse and/or select the terms by which records and/or data are described. This section provides examples for using indexes to:
1. Select the Nucleotide database.
2. Select Preview/Index.
3. Select the Properties index from the Preview/Index pull-down menu.
4. Leave the Preview/Index search box empty and select the 'Index' button to see an alphabetical listing of all index terms within the Properties index.
Index entries are listed alphabetically and Entrez will begin the index display at the very first entry (i.e., biomol genomic).
Use the scroll bar to view more entries. Use the Down and Up buttons to display the next set of entries in either direction. The Properties search field and its corresponding index are very useful. This field contains information about the GenBank division to which the record belongs (i.e., gbdiv inv). It also describes the molecule type and location. The Properties field also describes such things as whether the sequence is part of a population study or segmented set.
Compare the Properties index of the Nucleotide database to the Properties index of the other databases. A Properties index is not available for the Structure database.
Example: Examine the kind of information indexed by the Feature key index of the Genome database.
1. Select the Genome database.
2. Select Index.
3. Select the Feature key index from the View Index pull-down menu.
4. Type "0" (the number zero) without quotes in the View Index query box and select View.
Use the scroll bar to view the entries. Use the Up and Down buttons to display the next set of entries in either direction. The Feature key search field and its corresponding index are also very useful. This field contains information about the biological features of the nucleotide sequences as annotated by submitters and database staff.
Simply select EST database and enter your search terms in the search box.
1. Select the PopSet database.
2. Select Index.
3. Select the Organism index from the View Index pull-down menu.
4. Type "human" without quotes in the View Index query box and select View.
5. View the list of entries and locate the "human" entry.
6. Select the "human" entry by clicking on it once.
7. Select the "human" entry as a search term by clicking "AND". Note that the term is now located in the Search query box as "human" [Organism].
8. Type "mouse" without quotes in the View Index query box and select View.
9. View the list of entries and locate the "mouse" entry.
10. Select the "mouse" entry by clicking on it once.
11. Select the "mouse" entry as a search term by clicking "OR". Note that the term is now located in the Search query box with the human term (i.e., "human"[Organism] OR "mouse"[Organism]).
12. Repeat steps 8-11 above for Drosophila so that the final search statement in the query box is:
"human"[Organism] OR "mouse"[Organism] OR "drosophila"[Organism]
13. Select Go to execute this search.
1. Select the Protein database.
2. Select Index.
3. Select the Organism index from the View Index pull-down menu.
4. Type "pig" without quotes in the View Index query box and select View.
5. View the list of entries and locate the "pig" entry.
6. Select the "pig" entry by clicking on it once.
7. Select the "pig" entry as a search term by clicking "AND." Note that the term is now located in the Search query box as "pig" [Organism].
8. Select the Text Word index from the View Index pull-down menu.
9. Type "kinase" without quotes in the View Index query box and select View.
10. View the list of entries and locate the "kinase" entry.
11. Select the "kinase" entry by clicking on it once.
12. Select the "kinase" entry as a search term by clicking "AND". Note that the term is now located in the Search query box as "kinase" [Text Word] and that the final search statement in the query box is:
"pig"[Organism] AND "kinase"[Text Word]
13. Select Go to execute this search.
REMEMBER that Entrez processes complex search statements using Boolean Operators in a specific order as described in the Boolean Operators section above. You can always check the Details button to see how your final search statements were executed.
| Using Your History |
History provides a record of the searches performed during a search session. This section provides examples for using your search history to:
Please review how to use Boolean Operators.
1. Select the Protein database.
2. Type "streptomyces" in the query box and select Go.
3. Select Clear.
4. Type "pseudomonas" in the query box and select Go.
5. Select Clear.
6. Type "glucanase" in the query box and select Go.
7. Select History.
8. Review your search History and results. Note that each search statement is numbered. Also note the time and number of results for each search statement.
9. Combine the results of your earlier searches using the search numbers and Boolean operators. For example: ((#15) OR #16) AND #17. Select Go. Note: You can click on a "#" search result in 'History' to reveal an 'Options' menu.
10. Select History to once again review your search History and results.
Protein Database Glucanase Search History
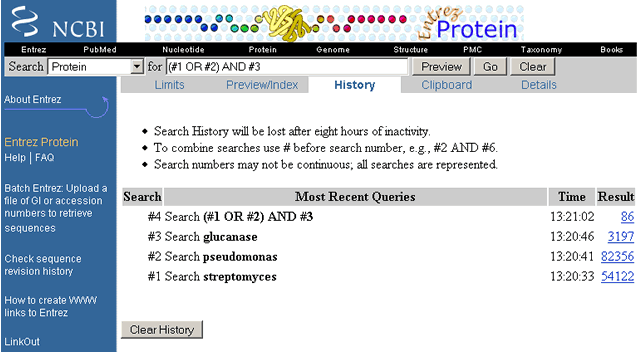
Although search Histories are database specific, the History numbering system is continuous across all databases searched during a single search session. For instance, let us say you just finished searching the Protein database using the example above. Next you want to search the Structure database for similar information. You cannot use your Protein database search History in the Structure database. However, as you start searching the Structure database, Entrez sequentially numbers the search sets based on the last search query executed in any database. Therefore, in this example, the first search query executed in the Structure database is numbered search #30. The next search query executed is numbered search #31 and so on. Entrez will save a maximum of 100 queries at a time.
A final note on search histories. If you search the same query in the same database during the same search session, the search set will only be saved in the History one time. To perform batch searches in Entrez, see or select Entrez Tools.
1. Select the Nucleotide database.
2. Type mouse[orgn] AND "fas antigen" with quotes around fas antigen in the query box and select Go.
3. The search retrieves over 30 documents. You do not want to review all the documents and decide you are really interested in any sequences with annotated exons or introns.
4. Select the 'History' tab.
5. Refine the results of your search using the search number and Boolean operators. For example: #1 AND (exon OR intron). Select 'Preview'.
6. Select 'History' to once again review your search History and results. Refining the search has reduced the number of retrieved documents to 9.
| Writing Advanced Search Statements |
Complex search statements can be written and executed directly from the the query box of any of the databases, as long as you obey some simple rules and use the correct syntax.
Perform a search by specifying the search terms, their fields, and the Boolean operations to perform on the term. Use the following syntax:
term [field] OPERATOR term [field]
Where term(s) are the search terms, the field(s) are the Search Fields and Qualifiers , and the OPERATOR(s) are the Boolean Operators. Remember that Boolean operators are normally processed from left to right. If you want part of your Boolean expression to be processed out of order, enclose it in parentheses.
Example: Find all human nucleotide sequences with D-loop annotations.
In the Nucleotide database, use the following expression:
D-loop[FKEY] AND human[ORGN]
Example: Find all human protein sequences with lengths between 50 and 60 amino acids that were entered into the database during 1999.
In the Protein database, use the following expression:
human[ORGN] AND 50[SLEN]:60[SLEN] AND 1999[MDAT]
Example: Find Drosophila population studies published in the Journal of Molecular Evolution
In the PopSet database, use the following expression:
j mol evol[JOUR] AND drosophila[ORGN]
| Displaying and Saving Results |
Entrez displays search results as shown below:
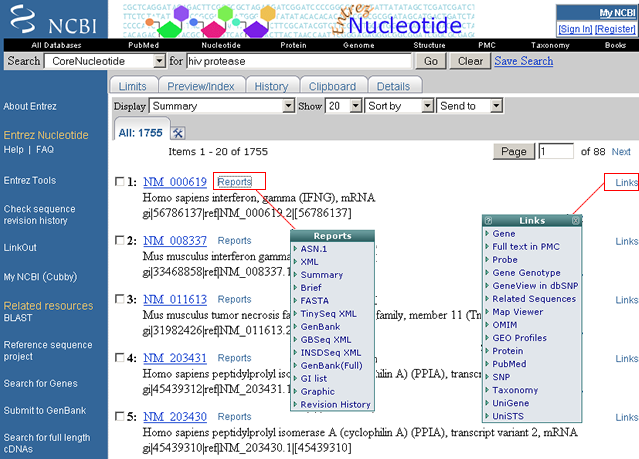
Document Summaries, or "docsums", are displayed for "hiv protease" search within the Nucleotide Database
The Search query box provides a summary of the database searched and the search terms as entered (i.e., "Search Nucleotide for hiv protease").
Clicking on the 'Links' button provides a dropdown list of databases that are linked to the individual document summary. Clicking on the 'Reports' button provides a dropdown list of data formats. The Sequence Revision History for the specific record can be viewed from the link 'Revision History'.
Sample Document Summary
| Display, Show, Sort, and Send to Options |
Display - The default display format is the Summary format shown in the example above.
To change the Display format, select an alternate format from the format pull-down menu (i.e., Summary). The format automatically displays.
To view the "graphical view", click on the accession number to display the GenBank report format. Select 'Graphics' from the Display menu. The Entrez graphical view will appear.
Show - The default number of documents displayed is 20. The total number of pages is displayed on the far right of the results page
To change the number of documents displayed per page, select an alternate number from the pull-down menu (e.g., 50) and, once selected, it will automatically display the new number selected.
In the Nucleotide database, for example, results can be sorted by accession number. Select the 'Sort by' pulldown menu and select the accession option. Send to - provides an option menu for sending the results to either text, file, or clipboard.
See the Display Formats table for a summary of the display formats available by database.
| Selecting Documents, Displaying Them, or Accessing Their Links |
Check boxes to the left of each document summary result are used to select individual documents from a set of documents retrieved. Once selected, the documents can be displayed (in various formats), sent to the Clipboard, or sent to a local disk. Documents are deselected by unclicking the check box.
Select Documents Using Check Boxes
For a useable FASTA format that can be easily used in other applications, select the Send to text option. The Send to text option uses your browser to display the sequence in FASTA format. Copy and paste the sequence from the browser to other applications. Also see the section below on saving to local disk for information on saving more useable data formats from Entrez.
| Details Button, Add To Clipboard, and Save |
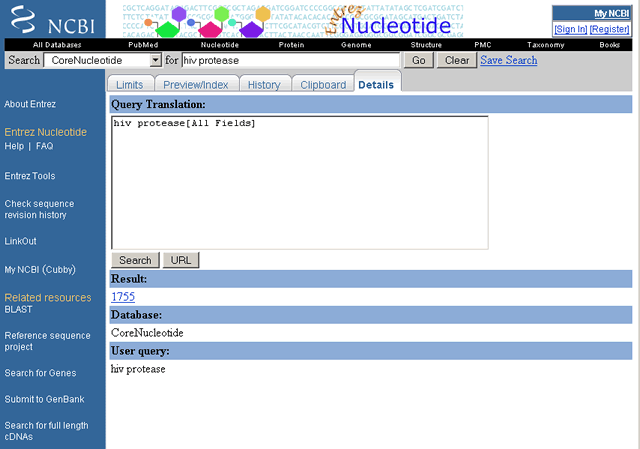
Details - click the Details tab to display your search strategy as translated using Entrez's search and syntax rules.
The Details window also contains error messages, when applicable. Note that the Details report shows the database searched, the number of documents retrieved (with hotlinks to the documents), and your search statement as written (i.e., not translated by Entrez). Within the Details window, you can modify and resubmit your search strategy. Submit the modified search query by selecting the Search button.
Details
Adding to the Clipboard - select documents 1, 3, and 5 from the results set by clicking on the check box adjacent to the document number. Then click the 'Clipboard' button. Note that three items were added to the Clipboard. You are also reminded that the Clipboard is limited to 500 items and that these three items will be lost after 8 hours of inactivity during a single search session. Also, please note that the document numbers for these items (i.e., documents 1, 3, and 5) are now shown in green to indicate that they are on the Clipboard. This feature is useful because as you continue to search, if these documents are retrieved through other search strategies, their document numbers will appear in green to indicate that they are already on the Clipboard.
Retrieving documents from the Clipboard - select the Clipboard button. The items on the Clipboard are displayed in the default Summary format. Note that the documents are renumbered, but the numbers are in green to indicate that the items are on the Clipboard. Also please note that you can display Clipboard items in all available formats, and you can link to document neighbors or related items in other databases. Items are removed from the Clipboard by selecting the items using the check box and selecting the “Clip Remove” option from the 'Send to' menu.
Saving to a local disk - select the Save button at the top (or bottom) of the results display screen next to the 'Text' button. Documents can also be saved from the Clipboard in the same manner described here. Before clicking the 'Save' button, decide two things: which documents you want and in what format. After selecting your documents by clicking on the check boxes and choosing the format using the format pull-down menu, select the 'Display' button. Once they are all displayed, click the Save button. You are prompted to name the file to which the results are saved on your local drive. If you do not select specific documents, all documents in the results set are saved. In the example below, documents 2, 3, 4, 6, and 9 will be saved to disk in the FASTA format. If these documents were not selected, all 30 documents (i.e., the entire retrieved set) would be saved to disk in the FASTA format.
Printing - use the Print function of your Web browser. As with saving to local disk, before printing, decide two things: Which documents you want to print and in what format. Because you are using the Web browser print function, you can only print documents that are displayed. Therefore, consider increasing the number of documents displayed per page so that the total number of documents you want to print are displayed on one page. Print hints: To save paper, consider using the Text or Save buttons before printing. Doing so will eliminate everything but the actual data you need (i.e., Entrez search interface, menu bars). If you use the Text button, print from your Web browser. If you use the Save button, print from another application on your machine.
| LinkOut |
LinkOut is a service that provides links from Entrez records to NCBI resources, such as UniGene and LocusLink, and to external resources, such as full-text journal articles, biological data, and sequence centers. These other resources provide a URL, resource name, and brief description of their Web site, which PubMed uses to create the links to their sites. User registration, a subscription fee, or some other type of fee may be required to access the full text of articles in some journals using this feature. Information for developers is available at: http://www.ncbi.nlm.nih.gov/entrez/linkout/doc/linkoutoverview.html