 Click
for full size image!
Click
for full size image! Click
for full size image!
Click
for full size image!
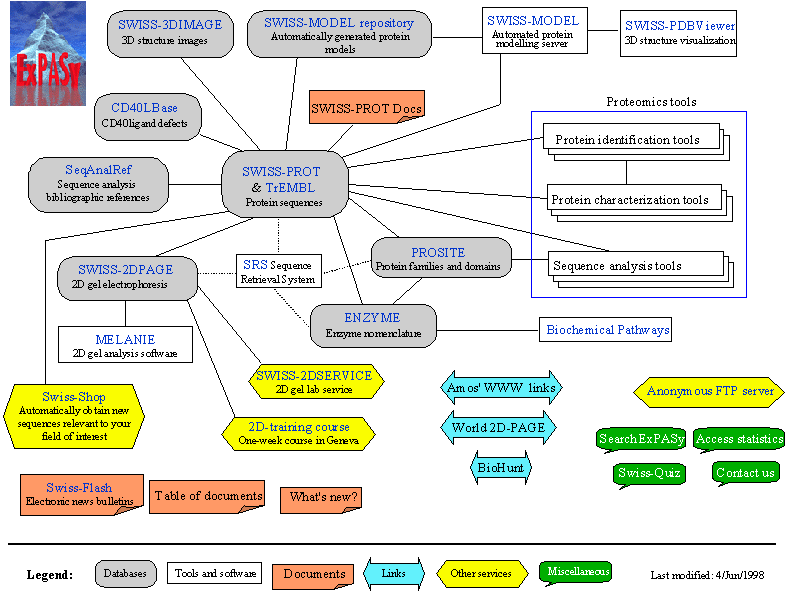
SwissProt and TrEMBL http://www.expasy.ch/sprot/
SWISS-PROT is a curated protein sequence database searched by
Keyword, Author, Title etc. It strives
to provide a high level of annotations (such as the
description of the function of a protein, its domains structure, post-translational
modifications, variants, etc.), a minimal level
of redundancy and high level of integration with other databases [More
details / References / Disclaimer].
TrEMBL is a computer-annotated supplement of SWISS-PROT that
contains all the translations of EMBL nucleotide
sequence entries not yet integrated in SWISS-PROT.
Output gives links to all records with hits to requested protein. Selection of a link brings up a NiceProt view of the data in that record. Links to literature refs; cross refs to other db's (PRODOM, DOMO, PROTOMAP, PFAM, others) ; structure and function info; features table; amino acid sequence.
PROSITE http://expasy.cbr.nrc.ca/prosite/
PROSITE is a database searched by protein families and domains. It consists of biologically significant sites, patterns and profiles that help to reliably identify to which known protein family (if any) a new sequence belongs.
Output is a NiceSite view of the data in SwissProt with
links to SWISS-PROT data entry for desired protein as well as related ones
with similar structure. Also links to PDB files and structural information.
Proteomics Tools http://www.expasy.ch/tools/#proteome
[Protein identification and characterization] [DNA -> Protein] [Similarity
searches] [Pattern and profile searches]
[Primary structure analysis] [Secondary structure prediction]
[Tertiary structure] [Transmembrane regions detection]
[Sequence alignment]