Complex I (NADH dehydrogenase)
Introduction:
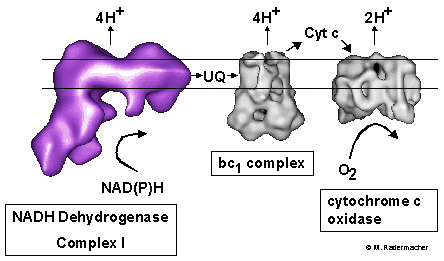
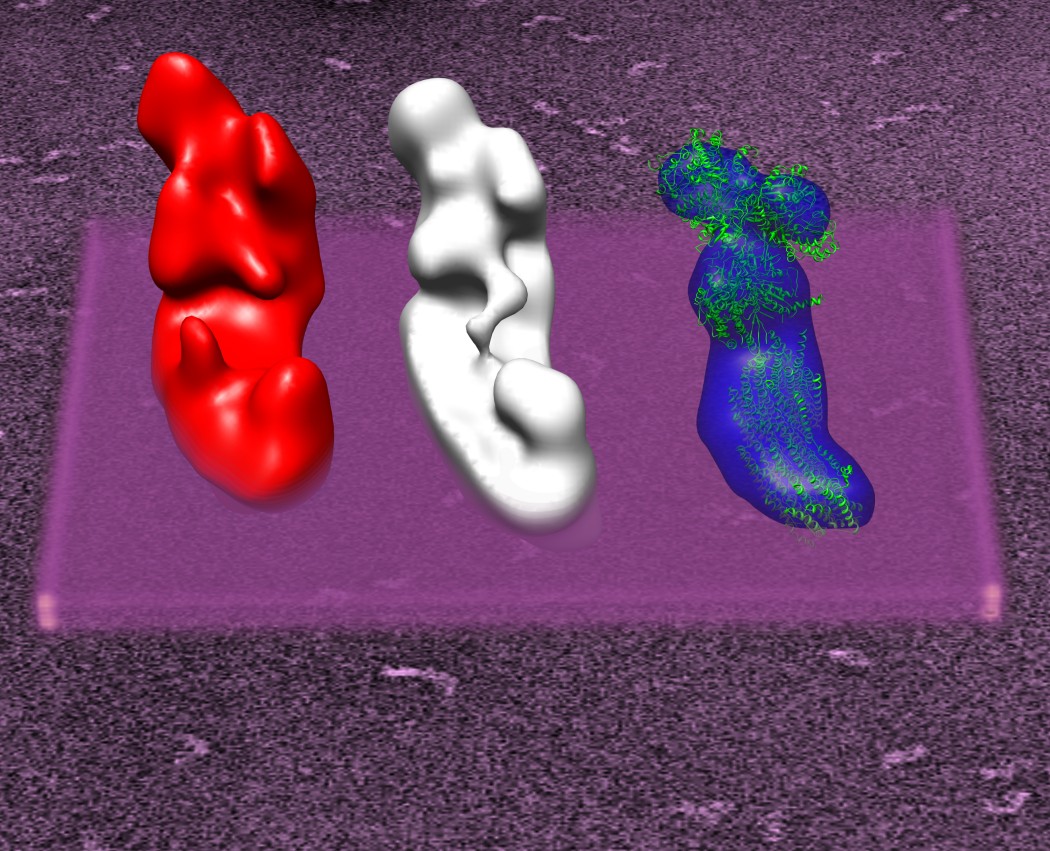
Structure and Function:

From 35 images one can construct 3×1038 movie sequences. We selected 15 images that show the central membrane arm protuberance moving from front to back/back to front and constructed by hand a highly speculative movie of possible conformational changes of complex I: Watch the movie
In some 2D averages of Y. lipolytica complex I and in some intermediate 3D reconstruction a faint line can be observed, that could indicate a very thin connection between the Matrix arm domain (5) and the membrane arm protrusions, assumed to be located near proton pumping subunits. This tether may be one way to transfer conformational changes in the Matrix arm to the membrane arm. For a conformationally driven complex I two pathways for conformational coupling may exist. Either through the tether, or through more internal conformational changes. The recent X-ray structures of complex I from E. coli and from T. thermophilus (Efremov et al. Nature, 2010, 465, 441-445, Baradaran et al. Nature 2013, 494,443-448) showed an unusual long horizontal helix in the membrane arm, that may be involved in the proton pumping mechanism.
The overall structure of complex I is preserved throughout species.
Selected Literature:
M. Radermacher, Chapter 1: Visualizing Functional Flexibility by Three-Dimensional Electron Microscopy: Reconstructing Complex I of the Mitochondrial Respiratory Chain. In: Methods in Enzymology, Mitochondrial Function, Part A: Mitochondrial Electron Transport Complexes and Reactive Oxygen Species, Edited by: William S. Allison and Immo E. Scheffler, (2009) Vol. 456, 3-27
Angerer H, Radermacher M, Małkowska M, Steger M, Zwicker K, Heide H, Wittig I, BrandtU, Zickermann V. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc Natl Acad Sci U S A. 2014 Apr 8;111(14):5207-12.
Radermacher M, Ruiz T, Clason T, Benjamin S, Brandt U, Zickermann V.The three-dimensional structure of complex I from Yarrowia lipolytica: a highly dynamic enzyme. J Struct Biol. 2006 Jun;154(3):269-79.
T. Clason, V. Zickermann, T. Ruiz, U. Brandt, M. Radermacher, Direct Localization of the 51 kDa and 24 kDa Subunits of Mitochondrial Complex I by Three-dimensional Difference Imaging, Journal of Structural Biology, (2007), 159(3):433-442.
Brandt U. Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem. 2006;75:69-92.
Abdrakhmanova A, Zickermann V, Bostina M, Radermacher M, Schagger H, Kerscher S, Brandt U.Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica.Biochim Biophys Acta. 2004 Jul 23;1658(1-2):148-56.
Zickermann V, Bostina M, Hunte C, Ruiz T, Radermacher M, Brandt U. Functional implications from an unexpected position of the 49-kDa subunit of NADH:ubiquinone oxidoreductase. J Biol Chem. 2003 Aug 1;278(31):29072-8.
Peng G, Fritzsch G, Zickermann V, Schagger H, Mentele R, Lottspeich F, Bostina M, Radermacher M, Huber R, Stetter KO, Michel H. Isolation, characterization and electron microscopic single particle analysis of the NADH:ubiquinone oxidoreductase (complex I) from the hyperthermophilic eubacterium Aquifex aeolicus. Biochemistry. 2003 Mar 18;42(10):3032-9.
Djafarzadeh R, Kerscher S, Zwicker K, Radermacher M, Lindahl M, Schagger H, Brandt U. Biophysical and structural characterization of proton-translocating NADH-dehydrogenase (complex I) from the strictly aerobic yeast Yarrowia lipolytica. Biochim Biophys Acta. 2000 Jul 20;1459(1):230-8.
Rasmusson AG, Heiser V V, Zabaleta E, Brennicke A, Grohmann L.Physiological, biochemical and molecular aspects of mitochondrial complex I in plants Biochim Biophys Acta. 1998 May 6;1364(2):101-11.