SOFTWARE
The immunogenomics data analysis working group is developing software intended to make the analysis of immunogenomic data simpler, faster, and more consistent across studies. Currently, the ANTT allele name translation application, the BIGDAWG R package and web-app for case-control analysis and the Global Frequency Map Browser are available for download or use over the internet.
| Bridging ImmunoGenomic Data-Analysis Workflow Gaps |
|
BIGDAWG is an integrated analysis system that automates the manual data-manipulation and trafficking steps (the gaps in an analysis workflow) normally required for analyses of highly polymorphic genetic systems (e.g., the HLA and KIR genes). BIGDAWG performs tests of Hardy-Weinberg equilibrium, and carries out case-control association analyses for haplotypes, individual loci, and HLA amino-acid positions on unambiguous genotype data. The 'BIGDAWG' R package (v1.3.4) is available for download from the Comprehensive R Archive Network (CRAN), and is available as an online web-app at bigdawg.immunogenomics.org. |
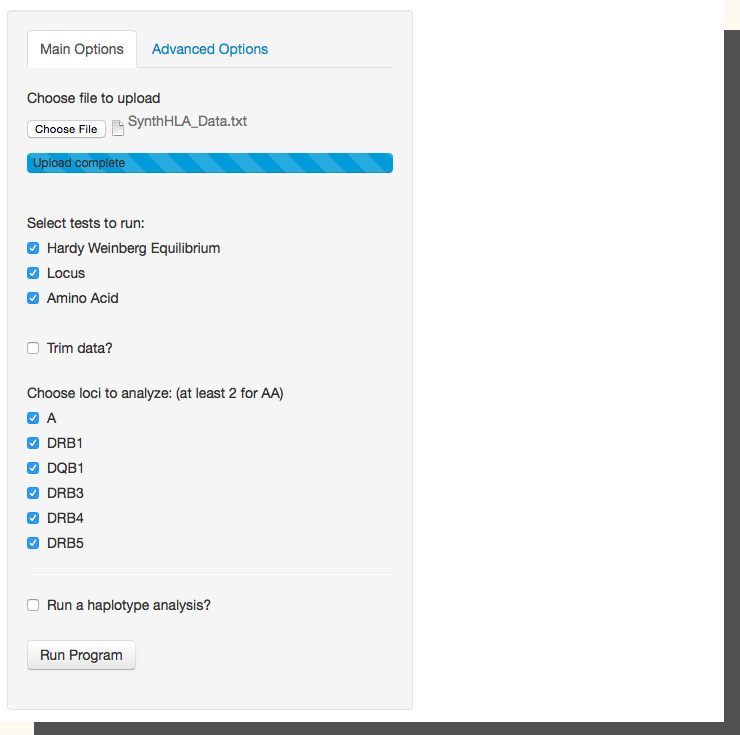
|

| Browser for Global Frequency Maps |
|
The Global Frequency Map Browser offers an easy way to view and compare the global HLA allele frequency maps at
www.pypop.org/popdata generated by Solberg et al. (2008).
|
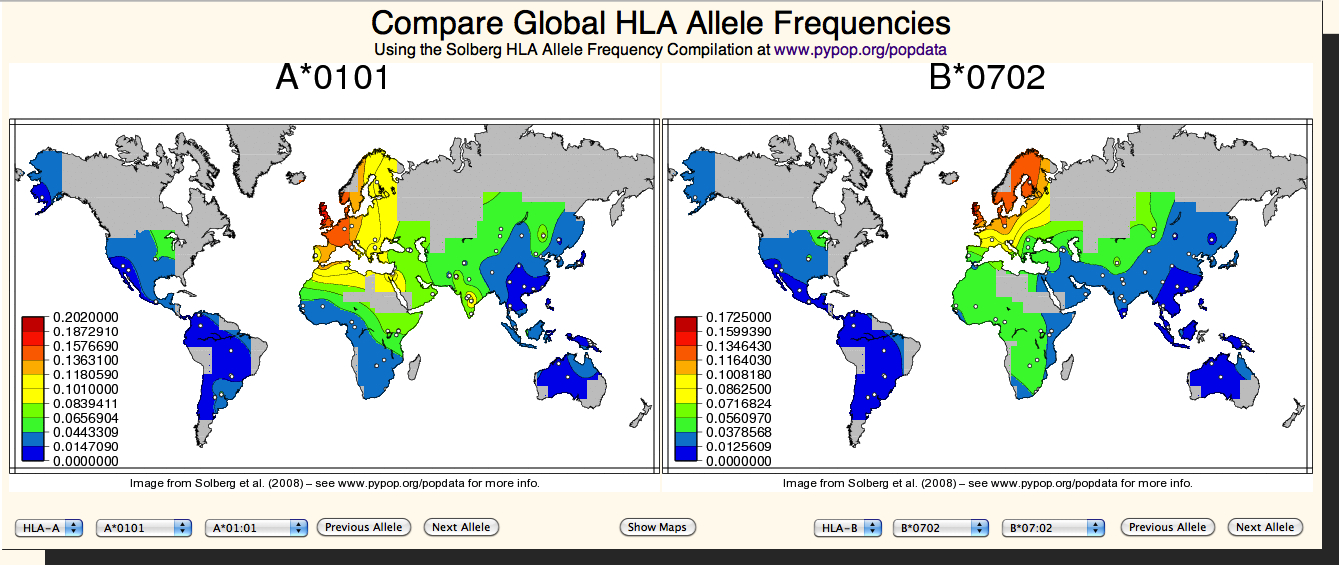
|
The GFM Browser was developed with the support of NIH grant U01AI067068.
|
Allele Name Translation
We have developed |
|
The Allele Name Translation Tool (ANTT) is a downloadable application that translates the allele names in entire datasets, and can be customized to translate allele names between any pair of naming conventions (inlcuding user-defined naming conventions).
December 11, 2010 ANTT version 0.5.0 is now available for use. |
|
Mack SJ, Hollenbach JA.
Allele Name Translation Tool and Update NomenCLature: Software tools for the automated translation of HLA allele names between successive nomenclatures.
Tissue Antigens 2010: 75(5):457-461.
DOI 10.1111/j.1399-0039.2010.01477.x
Acknowledgements: The ANTT was developed with the support of NIH grant U01AI067068.
We thank Glenys Thomson, Erik Sowa, Henry A. Erlich, Katrina R. Eaton, Jorge Oksenberg and Pierre-Antoine Gourraud, Pawinee Kupatawintu, Steven G. E. Marsh, James
Robinson and Hazael Maldonado Torres, Mike Varney and the Australian Red Cross Blood Service, Rachel Yedlin, Martin Maiers, and Thomas Smith for testing of the ANTT and UNCL
and helpful comments.