Gene assignment two hints:
As an example of how to find information about the mRNA and genomic sequence of your gene, here are some results for the Drosophila gene vermilion.
I went to the NCBI database and searched for "vermilion" in the Gene database.
Near the bottom of the page, I found a link to the mRNA sequence and to the genomic DNA sequence.
Here is the last part of the mRNA information: It shows that the mRNA transcript ("gene") is in position 1 to 1294 and that the protein coding sequence (CDS) starts at 39 and ends at 1178. If you go to position 39, you should find the start codon (ATG). Note that this sequence is a DNA copy of the mRNA, so the start codon (AUG in RNA) is ATG.

They also give the amino acid sequence for gene, as a string of one letter abbreviations of the amino acids.
The mature mRNA has already had the introns spliced out, and obviously does not include the promoter regions upstream of the transcription start. To see those you have to go to the genomic DNA sequence. Go back to the main gene page and follow links to the genomic sequence.
Fortunately, a lot of these genes have been annotated to show the positions of introns and exons. For example, here is part of the page corresponding to the vermilion genomic sequence:
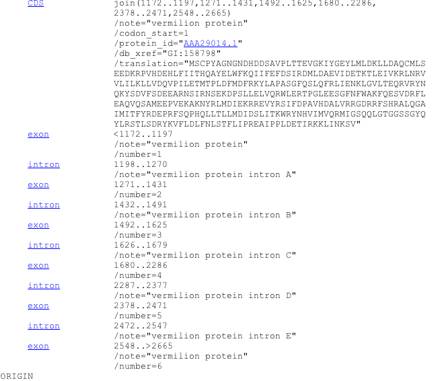
In this particular sequence, the first exon of the gene starts at 1172, then there is an intron that starts at 1198, a second exon at 1271, etc.
If your sequence has not been annotated to show introns, you might try searching for a different genomic sequence by going to the "nucleotide" database and searching for your gene. You may have to scan a long list to find a GENOMIC sequence that is from the same organism as your mRNA sequence.
Here is the beginning of the genomic DNA squence. You ought to be able to find the ATG that starts at position 1172, at the beginning of the first exon.
If you search for the first few bases of the mRNA (ccgctcga ...) you can find where that mRNA sequence starts. If the mRNA sequence is complete, that will be the beginning of transcription. In this case it is at 1135. (try it and make sure you can see see that those bases correspond to the beginning of the mRNA sequence.)

A few bases before 1135 you may be able to see some of the promoter sequences (e.g. TATA). The mRNA sequence in the database may not be the complete mRNA transcript, so the promoter sequences may be fairly far upstream. In this case I'm not sure where transcription starts, but there is a TATAAAA box at position 560, which may be near the start of the message.
However, finding promoters is fairly hard, because the sequences can vary. When I put this into a promoter search engine, it said the two most likely promoter sequences started at 165 or 290.
So, scan you gene and if you see something that looks likely, great. Otherwise don't get too frustrated abouit finding the promoters.
Finally, look for some of the intron sequences. For example the first intron begins with a GT at 1198 (as it should) and ends with an AG at 1270 (as it should).