Our Research
The topic of our continuing research is the development of new methods for 3D reconstruction of macromolecular assemblies and sub-cellular components. Methods we have developed include 3D reconstruction algorithms for conical tomography, the random conical reconstruction techniques combined with algorithms for 3D reconstruction from randomly oriented projections and techniques for reference based 3D reconstruction and refinements using Radon transform / polar Fourier transform based correlation methods. A fast reconstruction algorithm we developed is the two-step 3D Radon inversion algorithm. We have developed 3D multivariate statistical techniques applicable to volumes with missing data, for the analysis of conformational variations in 3D data sets.The technique uses the Probabilistic Principle Component Analysis with Expectation Maximization, where not only the eigenvectors are estimated but also the missing data. The technique allows for the restoration of single volumes with estimated missing data. Lately we have substantially improved non-linear mapping techniques, based on our original version published in 1985. Software developed in the course of our research is implemented in the package EMIRA (Environment for Modular Image Raconstruction Algorithms). For details and download see under the resources-tab.
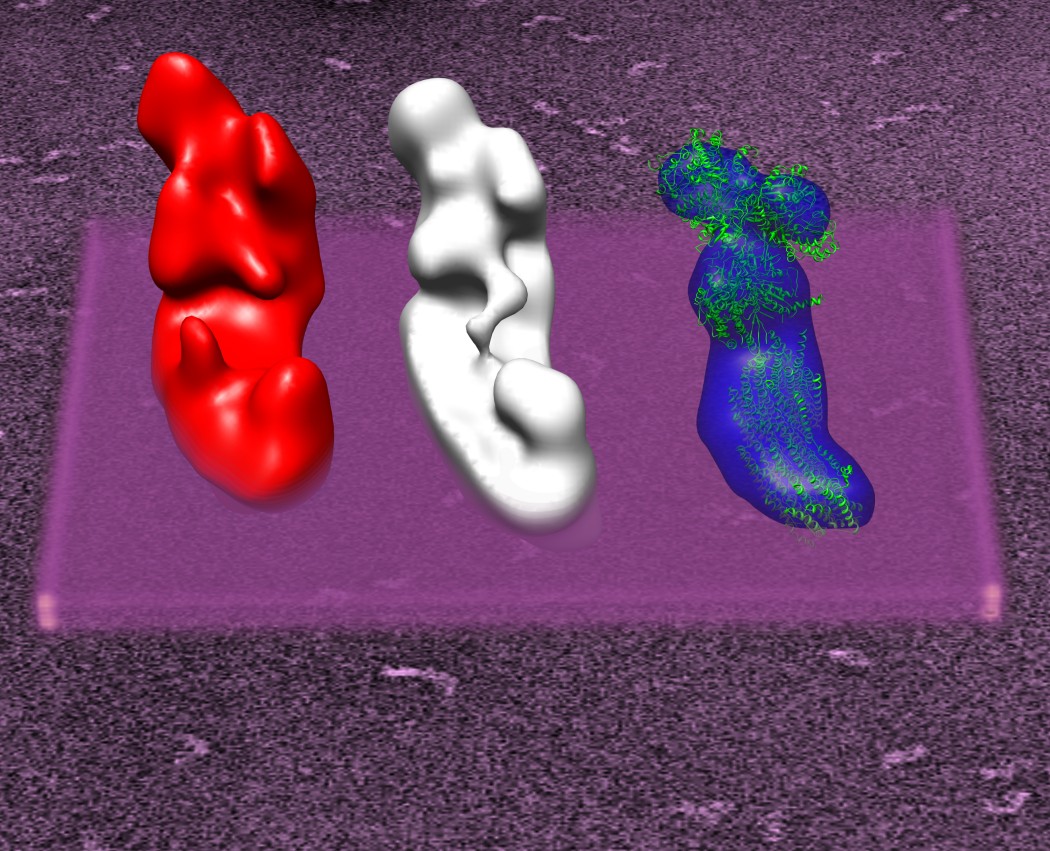
Our laboratory prviously has studied the structure and function of enzyme complexes in the mitochondrial respiratory chain by 3D electron microscopy. Our main emphasis is on the structure of complex I (NADH:ubiquinone oxidoreductase). Complex I is one of the entry points in the respiratory chain. It oxidizes NADH and in this process translocates protons across the inner mitochondrial membrane. Eukaryotic complex I has a molecular weight almost 1 MDa and is one of the largest membrane proteins in the mitochondrial inner membrane. It consists out of more than 40 individual subunits. 14 of the subunits are conserved throughout all species and are also found in bacterial complex I. We have shown, that not only are the central subunits conserved for all species but also its basic shape is the same for the yeast Yarrowia lipolytica , for bovine complex I and bacterial complex I from Aquifex aeolicus
New and selected structure determination projects are being carried out in collaboration with other laboratories.